Content from Introduction
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is this all about?
Objectives
- Explain the general structure of this introductory course
- Explain how to submit “homework” assignments
Introduction
This is the accompanying website to the Empra Differentielle Psychologie of Heidelberg University. It is authored and used by Sven Lesche. This course aims to cover:
- Some R basics and how to structure code
- Getting a first glimpse of data
- Cleaning data
- Running some analysis
Lesson Structure
The entire course is organized in short “Episodes” intended to take
around 10-15mins to read and around 5-10mins to complete the challenges
below. You will complete approximately one episode per week. At the end
of each episode, you will upload a script containing the solutions to
the challenges. The file format should be:
lessonnumber_solutions_name_surname.R. Upload the script
containing your solutions to a HeiBox folder and share that folder with
me.
A word about Chat-GPT
Chat-GPT is an amazing tool that makes it much easier to just try something out. Importantly, it can take a lot of work off your hands, especially early on in your learning process. Most of the challenges here can just be solved by plugging them into a LLM like Chat-GPT. I encourage you to first try to find a solution yourself. You will learn a lot more by failing yourself first and continuing to try.
Nonetheless, sometimes it may be necessary to ask Chat-GPT or google a solution. In real data analysis, this is done a lot! Nobody knows anything. However, the most important thing for now is that you understand the code you write. Thus, if you use Chat-GPT, make it explain to you why this code works and what it is doing or ask it for hints instead of the solution.
Accompanying Material
This course relies heavily on R for Data Science (2e), which can be treated as an accompanying textbook.
Questions & Support
This is the first version of the new course material. If you have any questions or notice any inconsistencies / opportunities for improvement, feel free to reach out to to me via mail or Slack. If you feel comfortable using GitHub, also feel free to submit an issue.
Some opportunities for improvement are:
- Is the material sufficiently explained? Can you follow?
- Are the exercises too difficult / not enough to practice material learned in an episode?
- Are you missing any information to complete following episodes?
- Is there information missing or information that you do not require?
- Are there any typos / wrong examples?
- Whatever else comes to mind.
Feel free to reach out to me via any of the channels with your feedback. I will try to implement it as soon as possible. Thank you for your help in making this course better for coming students!
- Follow the structured outline to learn R basics and data analysis
- Submit weekly scripts following the specified format
- Contact me via mail or Slack for any queries
Content from R Setup
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is R, what is R Studio?
- How to work with the R console and R scripts?
- What is an R package?
Objectives
- To gain familiarity with the various panes in the RStudio IDE
- To gain familiarity with the buttons, short cuts and options in the RStudio IDE
- To understand variables and how to assign to them
- To be able to manage your workspace in an interactive R session
- To be able to use mathematical and comparison operations
- To be able to call functions
- Introduction to package management
Introduction to RStudio
Welcome to the R portion of the Empra. This first lesson is adapted from resbaz’s introction to R workshop.
Throughout this lesson, you’re going to learn some of the fundamentals of the R language as well as some best practices for organizing code for scientific projects that will make your life easier.
We’ll be using RStudio: a free, open source R integrated development environment. It provides a built in editor, works on all platforms (including on servers) and provides many advantages such as integration with version control and project management.
Basic layout
When you first open RStudio, you will be greeted by three panels:
- The interactive R console (entire left)
- Environment/History (tabbed in upper right)
- Files/Plots/Packages/Help/Viewer (tabbed in lower right)
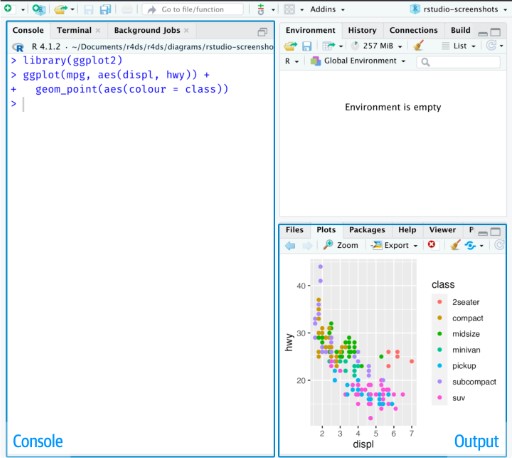
Once you open files, such as R scripts, an editor panel will also open in the top left.
Work flow within RStudio
There are two main ways one can work within RStudio.
- Test and play within the interactive R console then copy code into a
.R file to run later.
- This works well when doing small tests and initially starting off.
- It quickly becomes laborious
- Start writing in an .R file - called a script - and use RStudio’s
command / short cut to push current line, selected lines or modified
lines to the interactive R console.
- This is a great way to start; all your code is saved for later
- You will be able to run the file you create from within RStudio or
using R’s
source()function.
For now, let’s stick with the console. We will learn more about how to use R scripts later. Feel free to run all code examples provided here in your own RStudio console and figure out what they do.
Introduction to the R console
Much of your time in R will be spent in the R interactive console. This is where you will run all of your code, and can be a useful environment to try out ideas before adding them to an R script file.
The first thing you will see in the R interactive session is a bunch of information, followed by a “>” and a blinking cursor. R operates on the idea of a “Read, evaluate, print loop”: you type in commands, R tries to execute them, and then returns a result.
Using R as a calculator
The simplest thing you could do with R is do arithmetic:
R
1 + 100
OUTPUT
[1] 101And R will print out the answer, with a preceding “[1]”. Don’t worry about this for now, we’ll explain that later. For now think of it as indicating output.
If you type in an incomplete command, R will wait for you to complete it:
OUTPUT
+Any time you hit return and the R session shows a “+” instead of a “>”, it means it’s waiting for you to complete the command. If you want to cancel a command you can simply hit “Esc” and RStudio will give you back the “>” prompt.
Tip: Cancelling commands
If you’re using R from the commandline instead of from within
RStudio, you need to use Ctrl+C instead of Esc
to cancel the command. This applies to Mac users as well!
Cancelling a command isn’t just useful for killing incomplete commands: you can also use it to tell R to stop running code (for example if its taking much longer than you expect), or to get rid of the code you’re currently writing.
When using R as a calculator, the order of operations is the same as you would have learnt back in school.
From highest to lowest precedence:
- Parentheses:
(,) - Exponents:
^or** - Divide:
/ - Multiply:
* - Add:
+ - Subtract:
-
R
3 + 5 * 2
OUTPUT
[1] 13Use parentheses to group operations in order to force the order of evaluation if it differs from the default, or to make clear what you intend.
R
(3 + 5) * 2
OUTPUT
[1] 16This can get unwieldy when not needed, but clarifies your intentions. Remember that others may later read your code.
R
(3 + (5 * (2 ^ 2))) # hard to read
3 + 5 * 2 ^ 2 # clear, if you remember the rules
3 + 5 * (2 ^ 2) # if you forget some rules, this might help
The text after each line of code is called a “comment”. Anything that
follows after the hash (or octothorpe) symbol # is ignored
by R when it executes code.
Really small or large numbers get a scientific notation:
R
2/10000
OUTPUT
[1] 2e-04Which is shorthand for “multiplied by 10^XX”. So
2e-4 is shorthand for 2 * 10^(-4).
You can write numbers in scientific notation too:
R
5e3 # Note the lack of minus here
OUTPUT
[1] 5000Mathematical functions
R has many built in mathematical functions. To call a function, we simply type its name, followed by open and closing parentheses. Anything we type inside the parentheses is called the function’s arguments:
R
sin(1) # trigonometry functions
OUTPUT
[1] 0.841471R
log(1) # natural logarithm
OUTPUT
[1] 0R
log10(10) # base-10 logarithm
OUTPUT
[1] 1R
exp(0.5) # e^(1/2)
OUTPUT
[1] 1.648721Don’t worry about trying to remember every function in R. You can simply look them up on Google, or if you can remember the start of the function’s name, use the tab completion in RStudio.
This is one advantage that RStudio has over R on its own, it has autocompletion abilities that allow you to more easily look up functions, their arguments, and the values that they take.
Typing a ? before the name of a command will open the
help page for that command. As well as providing a detailed description
of the command and how it works, scrolling to the bottom of the help
page will usually show a collection of code examples which illustrate
command usage. Try reading the description to the log()
function by typing ?log() (or just ?log) in
the console.
Comparing things
Another useful feature of R next to functions are comparisons. Quite often, we want to see if one value is bigger than another or only use data with some particular value.
We can check if two values are equal by using the equality operator
==.
R
1 == 1 # equality (note two equals signs, read as "is equal to")
OUTPUT
[1] TRUER
1 != 2 # inequality (read as "is not equal to")
OUTPUT
[1] TRUER
1 < 0 # less than
OUTPUT
[1] FALSER
1 <= 1 # less than or equal to
OUTPUT
[1] TRUER
1 > 1 # greater than
OUTPUT
[1] FALSER
1 >= 1 # greater than or equal to
OUTPUT
[1] TRUEVariables and assignment
In the previous example, we simply used numbers to do comparisons.
However, most work in R will be done using variables. A
variable can be any quality, quantity or property we might be interested
in, like a response time, a score on a personality test or a diagnosis.
We can store values in variables using the assignment operator
<-, like this:
R
x <- 1/40
Notice that assignment does not print a value. Instead, we stored it
for later in something called a variable.
x now contains the value
0.025:
R
x
OUTPUT
[1] 0.025Look for the Environment tab in one of the panes of
RStudio, and you will see that x and its value have
appeared. Our variable x can be used in place of a number
in any calculation that expects a number:
R
log(x)
OUTPUT
[1] -3.688879Notice also that variables can be reassigned:
R
x <- 100
x used to contain the value 0.025 and and now it has the
value 100.
We can also update the value of a variable and store it with the same name again.
R
x <- x + 1 #notice how RStudio updates its description of x on the top right tab
The right hand side of the assignment can be any valid R expression.
The right hand side is fully evaluated before the assignment
occurs. This means that in the above example, x + 1 is
evaluated first and the result is only then assigned to the new
x.
Variable names can contain letters, numbers, underscores and periods. They cannot start with a number nor contain spaces at all. Different people use different conventions for long variable names, these include
- periods.between.words
- underscores_between_words
- camelCaseToSeparateWords
What you use is up to you, but be consistent.
R
this_is_okay
this.is.also.okay
someUseCamelCase
dont_be.aMenace_toSociety
I always use snake_case for variable names in R.
camelCase is often used in other programming languages such
as MATLAB or JavaScript.
Using = for assignment
It is also possible to use the = operator for
assignment:
R
x = 1/40
But this is much less common among R users. The most important thing
is to be consistent with the operator you use. There
are occasionally places where it is less confusing to use
<- than =, and it is the most common symbol
used in the community. So the recommendation is to use
<-.
If we try to give a variable an invalid name, R will throw an error.
ERROR
Error in parse(text = input): <text>:1:2: unexpected input
1: 2_
^Tip: Warnings vs. Errors
Pay attention when R does something unexpected! Errors, like above, are thrown when R cannot proceed with a calculation. Warnings on the other hand usually mean that the function has run, but it probably hasn’t worked as expected.
In both cases, the message that R prints out usually give you clues how to fix a problem.
Errors can be frustrating, but they are your friend! They tell you that something went wrong and usually give you an informative message as to what went wrong. If you cannot make something of the error message, try pasting it into Google or Chat-GPT, this will often help.
Challenges
Challenge 2
What will be the value of each variable after each statement in the following program? You are encouraged to run these commands in R.
R
mass <- 47.5
age <- 122
mass <- mass * 2.3
age <- age - 20
Challenge 3
Run the code from the previous challenge, and write a command to compare mass to age. Is mass larger than age?
- R Studio is a shiny environment that helps you write R code
- You can either write code directly in the console, or use script to organize and save your code
- You can assign variables using
<- - Be consistent in naming variables, other people should be able to read and understand your code
Content from Packages in R
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is an R package?
- How can we use R packages
Objectives
- Explain how to use R packages
- Explain how to install R packages
- Understand the difference between installing and loading an R package
R Packages
Per default, R provides you with some basic functions like
sum(), mean() or t.test(). These
functions can already accomplish a lot, but for more specialized
analyses or more user-friendly functions, you might want to use
additional functions.
If you are in need of a specific function to achieve your goal, you can either write it yourself (more on this later) or use functions written by other users. These functions are often collected in so-called “packages”. The official source for these packages on R is CRAN (the comprehensive R archive network).
Packages you may encounter
Packages make R really powerful. For 95% of your analysis-needs,
there probably exists a package designed specifically to hand this. For
example, some packages you might use often are tidyverse
for data cleaning, psych for some psychology specific
functions, afex for ANOVAs or lme4 for
multi-level models. You can even use R packages for more complicated
analyses like structural equation models (lavaan) or
bayesian modeling (brms). You can even write papers using R
using papaya. Even this website was written using the
R-packages rmarkdown and sandpaper.
CRAN makes it really easy to use the over 7000 R packages other users
provide. You can install them using
install.packages("packagename") with the name of the
package in quotation marks. This installs all functionalities of this
packages on your machine. However, this package is not automatically
available to you. Before using it in a script (or the console) you need
to tell R to “activate” this package. You can do this using
library(packagename). This avoids loading all installed
packages every time R is starting (which would take a while).
Using functions without loading a package
If you are only using a few functions from a certain package (maybe
even only once), you can avoid loading the entire package and only
specifically access that function using the :: operator.
You can do this by typing packagename::function(). If the
package is installed, it will allow you to use that function without
calling library(packagename) first. This may also be useful
in cases where you want to allow the reader of your code to easily
understand what package you used for a certain function.
Demonstration
First, we need to install a package. This will often generate a lot of text in your console. This is nothing to worry about. In most cases, it is enough to look at the last few messages, they will tell you what went wrong or whether everything went right.
R
install.packages("dplyr")
OUTPUT
The following package(s) will be installed:
- dplyr [1.1.4]
These packages will be installed into "~/work/r-for-empra/r-for-empra/renv/profiles/lesson-requirements/renv/library/linux-ubuntu-jammy/R-4.5/x86_64-pc-linux-gnu".
# Installing packages --------------------------------------------------------
- Installing dplyr ... OK [linked from cache]
Successfully installed 1 package in 5 milliseconds.Then, we will need to load the package to make its functions
available for use. For most packages, this will also print a lot of
messages in the console in the bottom left. Again, this is usually
harmless. If something does go wrong, you will see the word
Error: along with a message somewhere.
Warnings: can often be ignored in package installation.
R
# Loading the package dplyr
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionNow we can use all the functions that dplyr provides.
Let’s start by using glimpse() to get a quick glance at
some data. For this case, we are using the iris data, that
comes with your default R installation.
R
# iris is a dataset provided as default in R
glimpse(iris)
OUTPUT
Rows: 150
Columns: 5
$ Sepal.Length <dbl> 5.1, 4.9, 4.7, 4.6, 5.0, 5.4, 4.6, 5.0, 4.4, 4.9, 5.4, 4.…
$ Sepal.Width <dbl> 3.5, 3.0, 3.2, 3.1, 3.6, 3.9, 3.4, 3.4, 2.9, 3.1, 3.7, 3.…
$ Petal.Length <dbl> 1.4, 1.4, 1.3, 1.5, 1.4, 1.7, 1.4, 1.5, 1.4, 1.5, 1.5, 1.…
$ Petal.Width <dbl> 0.2, 0.2, 0.2, 0.2, 0.2, 0.4, 0.3, 0.2, 0.2, 0.1, 0.2, 0.…
$ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, setosa, s…R
# Using a function without loading the entire package
# dplyr::glimpse(iris)
Here, we can see that iris has 150 rows (or
observations) and 5 columns (or variables). The first four variables are
size measurements regarding length and width of sepal and petal and the
fifth variable is a variable containing the species of flower.
Challenges
Challenge 1:
Install the following packages: dplyr,
ggplot2, and psych.
Challenge 2:
Load the package dplyr and get an overview of the data
mtcars using glimpse().
Challenge 3:
Figure out what kind of data mtcars contains. Make a
list of the columns in the dataset and what they might mean.
Hint
You are allowed to use Google (or other sources) for this. It is common practice to google information you don’t know or look online for code that might help.
Challenge 4:
Use the function describe() from the package
psych without loading it first.
What differences do you notice between glimpse() and
describe()?
- R packages are “add-ons” to R, they provide useful new tools.
- Install a package using
install.packages("packagename"). - Use a package using
library(packagename)at the beginning of your script. - Use
::to access specific functions from a package without loading it entirely.
Content from Vectors and variable types
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- How do you use scripts in R to document your work?
- How can you organize scripts effectively?
- What is a vector?
Objectives
- Show how to use scripts to generate reproducible analyses
- Explain what a vector is
- Explain variable types
- Show examples of
sum()andmean()
Scripts in R
For now, we have been only using the R console to execute commands. This works great if there are some quick calculations you have to run, but is unsuited for more complex analyses. If you want to reproduce the exact steps you took in data cleaning and analyses, you need to write them down - like a recipe.
This is where R scripts come in. They are basically like a text file (similar to Word) where you can write down all the steps you took. This makes it possible to retrace them and produce the exact same result over and over again.
In order to use R scripts effectively, you need to do two things.
Firstly, you need to write them in a way that is understandable in the
future. We will learn more about how to write clean code in future
lessons. Secondly (and maybe more importantly) you need to actually
save these scripts on your computer, and ideally save
them in a place where you can find them again. The place where you save
your script and especially the place you save your data should ideally
be a folder in a sensible place. For example, this script is saved in a
sub-folder episodes/ of the workshop folder
r_for_empra/. This makes it easy for humans to find the
script. Aim for similar clarity in your folder structure!
For this lesson, create a folder called r_for_empra
somewhere on your computer where it makes sense for you. Then, create a
new R script by clicking File > New File > R Script
or hitting Ctrl + Shift + N. Save this script as
episode_scripts_and_vectors.R in the folder
r_for_empra created above. Follow along with the episode
and note down the commands in your script.
Using a script
Let’s start learning about some basic elements of R programming. We
have already tried assigning values to variables using the
<- operator. For example, we might assign the constant
km_to_m the value 1000.
R
km_to_m <- 1000
Now, if we had data on distances in km, we could also use this value to compute the distance in meters.
R
distance_a_to_b_km <- 7.56
distance_a_to_b_m <- distance_a_to_b_km * km_to_m
If we have multiple distances and want to transform them from km to m
at the same time, we can make use of a vector. A vector is just a
collection of elements. We can create a vector using the function
c() (for combine).
Tip: Running segments of your code
RStudio offers you great flexibility in running code from within the
editor window. There are buttons, menu choices, and keyboard shortcuts.
To run the current line, you can 1. click on the Run button
just above the editor panel, or 2. select “Run Lines” from the “Code”
menu, or 3. hit Ctrl + Enter in Windows or Linux or
Command-Enter on OS X. (This shortcut can also be seen by hovering the
mouse over the button). To run a block of code, select it and then
Run via the button or the keyboard-shortcut
Ctrl + Enter.
Variable Types
Vectors can only contain values of the same type. There are two basic types of variables that you will have to interact with - numeric and character variables. Numeric variables are any numbers, character variables are bits of text.
R
# These are numeric variables:
vector_of_numeric_variables <- c(4, 7.642, 9e5, 1/97) # recall, 9e5 = 9*10^5
# Show the output
vector_of_numeric_variables
OUTPUT
[1] 4.000000e+00 7.642000e+00 9.000000e+05 1.030928e-02R
# These are character variables:
vector_of_character_variables <- c("This is a character variable", "A second variable", "And another one")
# Show the output
vector_of_character_variables
OUTPUT
[1] "This is a character variable" "A second variable"
[3] "And another one" We can not only combine single elements into a vector, but also combine multiple vectors into one long vector.
R
numeric_vector_1 <- c(1, 2, 3, 4, 5)
numeric_vector_2 <- c(6:10) # 6:10 generates the values 6, 7, 8, 9, 10
combined_vector <- c(numeric_vector_1, numeric_vector_2)
combined_vector
OUTPUT
[1] 1 2 3 4 5 6 7 8 9 10Recall that all elements have to be of the same type. If you try to combine numeric vectors with character vectors, R will automatically convert everything to a character vector (as this has less restrictions, anything can be a character).
R
character_vector_1 <- c("6", "7", "8")
# Note that the numbers are now in quotation marks!
# They will be treated as characters, not numerics!
combining_numeric_and_character <- c(numeric_vector_1, character_vector_1)
combining_numeric_and_character
OUTPUT
[1] "1" "2" "3" "4" "5" "6" "7" "8"We can fix this issue by converting the vectors to the same type. Note how the characters are also just numbers, but in quotation marks. Sometimes, this happens in real data, too. Some programs store every variable as a character (sometimes also called string). We then have to convert the numbers back to the number format:
R
converted_character_vector_1 <- as.numeric(character_vector_1)
combining_numeric_and_converted_character <- c(numeric_vector_1, converted_character_vector_1)
combining_numeric_and_converted_character
OUTPUT
[1] 1 2 3 4 5 6 7 8But be careful, this conversion does not always work! If R does not
know how to convert a specific character to a number, it will simply
replace this with NA.
R
character_vector_2 <- c("10", "11", "text")
# The value "text", can not be interpreted as a number
as.numeric(character_vector_2)
WARNING
Warning: NAs introduced by coercionOUTPUT
[1] 10 11 NAInspecting the type of a variable
You can use the function str() to learn about the
structure of a variable. The first entry of the output tells us about
the type of the variable.
R
str(vector_of_numeric_variables)
OUTPUT
num [1:4] 4.00 7.64 9.00e+05 1.03e-02This tells us that there is a numeric vector, hence num, with 4 elements.
R
str(vector_of_character_variables)
OUTPUT
chr [1:3] "This is a character variable" "A second variable" ...This tells us that there is a character vector, hence chr, with 3 elements.
R
str(1:5)
OUTPUT
int [1:5] 1 2 3 4 5Note that this prints int and not the usual num. This is because the vector only contains integers, so whole numbers. These are stored in a special type that takes up less memory, because the numbers need to be stored with less precision. You can treat it as very similar to a numeric vector, and do all the same wonderful things with it!
Simple functions for vectors
Let’s use something more exciting than a sequence from 1 to 10 as an
example vector. Here, we use the mtcars data that you
already got to know in an earlier lesson. mtcars carries
information about cars, like their name, fuel usage, weight, etc. This
information is stored in a data frame. A data frame is a
rectangular collection of variables (in the columns) and observations
(in the rows). In order to extract vectors from a data frame we can use
the $ operator. data$column extracts a
vector.
R
mtcars_weight_tons <- mtcars$wt
# note that it is a good idea to include the unit in the variable name
mtcars_weight_tons
OUTPUT
[1] 2.620 2.875 2.320 3.215 3.440 3.460 3.570 3.190 3.150 3.440 3.440 4.070
[13] 3.730 3.780 5.250 5.424 5.345 2.200 1.615 1.835 2.465 3.520 3.435 3.840
[25] 3.845 1.935 2.140 1.513 3.170 2.770 3.570 2.780Let’s start learning some basic information about this vector:
R
str(mtcars_weight_tons)
OUTPUT
num [1:32] 2.62 2.88 2.32 3.21 3.44 ...The vector is of type numeric and contains 32 entries. We can double
check this by looking into our environment tab, where
num [1:32] indicates just that. Similarly, we can get the
length of a vector by using length()
R
length(mtcars_weight_tons)
OUTPUT
[1] 32We can compute some basic descriptive information of the weight of
cars in the mtcars data using base-R functions:
R
# Mean weight:
mean(mtcars_weight_tons)
OUTPUT
[1] 3.21725R
# Median weight:
median(mtcars_weight_tons)
OUTPUT
[1] 3.325R
# Standard deviation:
sd(mtcars_weight_tons)
OUTPUT
[1] 0.9784574To get the minimum and maximum value, we can use min()
and max().
We can also get a quick overview of the weight distribution using
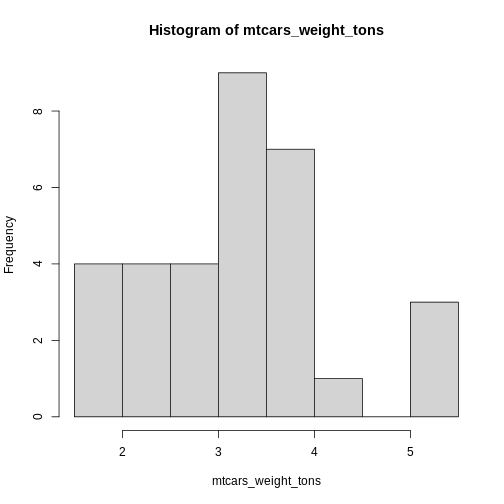
hist(), which generates a simple histogram.
R
hist(mtcars_weight_tons)
Histograms
Histograms are more powerful tools than it seems on first glance. They allow you to simply gather knowledge about the distribution of your data. This is especially important for us psychologists. Do we have ceiling effects in the task? Were some response options even used? How is response time distributed?
If your histogram is not detailed enough, try adjusting the
breaks parameter. This tells hist() how many
bars to print in the plot.
R
hist(mtcars_weight_tons, breaks = 30)
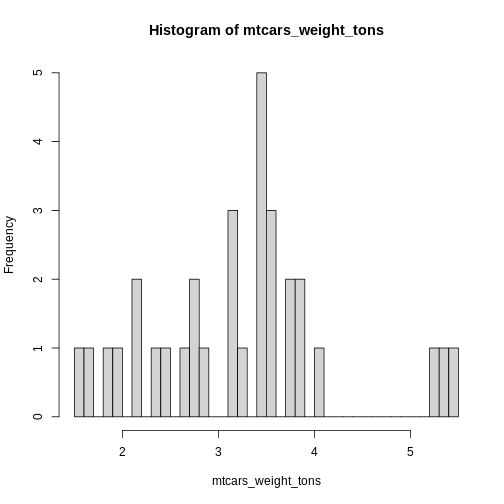
The golden rule for the number of breaks is simple: try it until it looks good! You are free to explore here.
Another useful function is unique(). This function
removes duplicates and only returns unique values. I use it a lot in
experimental data. Since every participant contributes multiple trials,
but I am sometimes interested in values a participant only contributes
once, I can use unique() to only retain each value
once.
In mtcars data, we might want to see how many cylinders
are possible in the data. unique(mtcars$cyl) is much easier
to read at a glance.
R
mtcars$cyl
OUTPUT
[1] 6 6 4 6 8 6 8 4 4 6 6 8 8 8 8 8 8 4 4 4 4 8 8 8 8 4 4 4 8 6 8 4R
unique(mtcars$cyl)
OUTPUT
[1] 6 4 8Tip: Unique + Length = Quick Count
Combining unique() and length() leads to a
really useful property. This returns the number of unique entries in a
vector. I use it almost daily! For example to figure out how many
participants are in a given data frame.
unique(data$participant_id) returns all the different
participant ids and length(unique(data$participant_id))
then gives me the number of different ids, so the number of participants
in my data.
Indexing and slicing vectors
There is one more important thing to learn about vectors before we move on to more complicated data structures. Most often, you are going to use the whole vector for computation. But sometimes you may only want the first 150 entries, or only entries belonging to a certain group. Here, you must be able to access specific elements of the vector.
In the simplest case, you can access the \(n\)th element in a vector by using
vector[n]. To access the first entry, use
vector[1], and so on.
R
test_indexing_vector <- seq(1, 32, 1)
# Seq generates a sequence from the first argument (1) to the second argument (32)
# The size of the steps is given by the third argument
test_indexing_vector
OUTPUT
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
[26] 26 27 28 29 30 31 32R
test_indexing_vector[1]
OUTPUT
[1] 1R
test_indexing_vector[c(1, 2)]
OUTPUT
[1] 1 2You can also remove an element (or multiple) from a vector by using a
negative sign -.
R
test_indexing_vector[-c(11:32)] # this removes all indexes from 11 to 32
OUTPUT
[1] 1 2 3 4 5 6 7 8 9 10The same rules apply for all types of vectors (numeric and character).
Tip: Think about the future
Excluding by index is a simple way to clean vectors. However, think
about what happens if you accidentally run the command
vector <- vector[-1] twice, instead of once?
The first element is going to be excluded twice. This means that the vector will lose two elements. In principle, it is a good idea to write code that you can run multiple times without causing unwanted issues. To achieve this, either use a different way to exclude the first element that we will talk about later, or simply assign the cleaned vector a new name:
R
cleaned_vector <- vector[-1]
Now, no matter how often you run this piece of code,
cleaned_vector will always be vector without
the first entry.
Filtering vectors
In most cases, you don’t know what the element of the vector is you want to exclude. You might know that some values are impossible or not of interest, but don’t know where they are. For example, the accuracy vector of a response time task might look like this:
R
accuracy_coded <- c(0, 1, 1, 1, 1, -2, 99, 1, 11, 0)
accuracy_coded
OUTPUT
[1] 0 1 1 1 1 -2 99 1 11 0-2 and 99 are often used to indicate
invalid button-presses or missing responses. 11 in this
case is a wrong entry that should be corrected to 1.
If we were to compute the average accuracy now using
mean() we would receive a wrong response.
R
mean(accuracy_coded)
OUTPUT
[1] 11.3Therefore, we need to exclude all invalid values before continuing our analysis. Before I show you how I would go about doing this, think about it yourself. You do not need to know the code, yet. Just think about some rules and steps that need to be taken to clean up the accuracy vector. What comparisons could you use?
Important: The silent ones are the worst
The above example illustrates something very important. R will not
throw an error every time you do something that doesn’t make sense. You
should be really careful of “silent” errors. The mean()
function above works exactly as intended, but returns a completely
nonsensical value. You should always conduct sanity checks. Can
mean accuracy be higher than 1? How many entries am I expecting in my
data?
Now, back to the solution to the wacky accuracy data. Note that R gives us the opportunity to do things in a million different ways. If you came up with something different from what was presented here, great! Make sure it works and if so, use it!
First, we need to recode the wrong entry. That 11 was
supposed to be a 1, but someone entered the wrong data in
the excel. To do this, we can find the index, or the element number,
where accuracy is 11. Then, we can replace that entry with 1.
R
index_where_accuracy_11 <- which(accuracy_coded == 11)
accuracy_coded[index_where_accuracy_11] <- 1
# or in one line:
# accuracy_coded[which(accuracy_coded == 11)] <- 1
accuracy_coded
OUTPUT
[1] 0 1 1 1 1 -2 99 1 1 0Now, we can simply exclude all values that are not equal to 1 or 0.
We do this using the - operators:
R
accuracy_coded[-which(accuracy_coded != 0 & accuracy_coded != 1)]
OUTPUT
[1] 0 1 1 1 1 1 1 0However, note that this reduces the number of entries we have in our
vector. This may not always be advisable. Therefore, it is often better
to replace invalid values with NA. The value
NA (not available) indicates that something is not a
number, but just missing.
R
# Note that now we are not using the - operator
# We want to replace values that are not 0 or 1
accuracy_coded[which(accuracy_coded != 0 & accuracy_coded != 1)] <- NA
Now, we can finally compute the average accuracy in our fictional experiment.
R
mean(accuracy_coded)
OUTPUT
[1] NAChallenges
Challenge 1:
We did not get a number in the above code. Figure out why and how to fix this. You are encourage to seek help on the internet.
Challenge 2:
Below is a vector of response times. Compute the mean response time, the standard deviation, and get the number of entries in this vector.
R
response_times_ms <- c(
230.7298, 620.6292, 188.8168, 926.2940, 887.4730,
868.6299, 834.5548, 875.2154, 239.7057, 667.3095,
-142.891, 10000, 876.9879
)
Challenge 3:
There might be some wrong values in the response times vector from
above. Use hist() to inspect the distribution of response
times. Figure out which values are implausible and exclude them.
Recompute the mean response time, standard deviation, and the number of
entries.
Challenge 4:
Get the average weight (column wt) of cars in the
mtcars data . Can you spot any outliers in the
histogram?
Exclude the “outlier” and rerun the analyses.
Challenge 5:
Get the mean values of responses to this fictional questionnaire:
R
item_15_responses <- c("1", "3", "2", "1", "2", "none", "4", "1", "3", "2", "1", "1", "none")
Challenge 6 (difficult):
Compute the average of responses that are valid as indicated by the
vector is_response_valid:
R
response_values <- c(91, 82, 69, 74, 62, 19, 84, 61, 92, 53)
is_response_valid <- c("yes", "yes", "yes", "yes", "yes",
"no", "yes", "yes", "yes", "no")
R
valid_responses <- response_values[which(is_response_valid == "yes")]
mean(valid_responses)
OUTPUT
[1] 76.875- Scripts facilitate reproducible research
- create vectors using
c() - Numeric variables are used for computations, character variables often contain additional information
- You can index vectors by using
vector[index]to return or exclude specific indices - Use
which()to filter vectors based on specific conditions
Content from Projects
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is an R project?
- How do projects help me organize my scripts.
Objectives
- Explain how projects can provide structure
- Explain what a “working directory” is and how to access files
Working Directory
Last week we worked with scripts and some basic computations using vectors. Most likely, you wrote down a lot of code and saved the script somewhere on your computer. Once you start working with multiple scripts and datasets, it becomes very important that you keep your files somewhat orderly.
This is important both for us humans to understand, but also for computers. Any time you need to access something outside your present R script, you will need to tell R where to find this. The data will be saved somewhere on the computer and needs to find its way into the R session.
Understanding where a file is saved on your computer is key to understanding how to tell R to read it into your current session. There are two main ways to tell R how to find a file: absolute paths and relative paths.
Absolute Paths
An absolute path is the full location of a file on your computer, starting from the root of your file system. It tells R exactly where to find the file, no matter where your R script is located. Think of it like an address in a town. 1404 Mansfield St, Chippewa Falls, WI 54729 tells you exactly where a house is located.
For example, the absolute path to a file stored in the
Documents/ folder most likely looks something like
this:
-
Windows:
"C:/Users/YourName/Documents/my_data.csv"
-
Mac/Linux:
"/Users/YourName/Documents/my_data.csv"
How to Find the Absolute Path of a File:
On Windows, open File Explorer, navigate to your file, right-click it, and select Properties. The full file path will be shown in the “Location” field.
On Mac, open Finder, right-click the file, select Get Info, and look at “Where.”
Relative Paths
A relative path specifies the location of a file in relation to the working directory of your R session. This is useful when working with projects that contain multiple files, as it keeps your code flexible and portable. Think of this like the direction, “just follow the road until the roundabout and then take the second exit”. If the starting point is well defined, this is enough information to find the target.
The working directory is the default folder on your computer where R looks for files and saves new ones. Think of it as R’s “home base”—if you ask R to read or save a file without giving a full path, it will assume you’re talking about this location.
You can check your current working directory by running:
R
getwd() # This tells you where R is currently looking for files
OUTPUT
[1] "/home/runner/work/r-for-empra/r-for-empra/site/built"In our case, this means if you try to read a file like this:
R
read.csv("data.csv")
R will look for data.csv inside
/home/runner/work/r-for-empra/r-for-empra/site/built.
If your files are stored somewhere else, you can change the working directory using:
R
setwd("C:/Users/YourName/Desktop/MyNewFolder") # Set a new working directory
Now, R will assume all file paths start from
"C:/Users/YourName/Desktop/MyNewFolder".
There are two main ways to define the working directory that R will
use. You can do this using setwd() and specify a particular
directory you want to start from. Another way to accomplish this is
through the use of R projects. These projects
automatically set your working directory to the place that the project
is located. More about projects in just a second.
If you prefer declaring your working directory using
setwd(), you can place this bit of code
setwd(dirname(rstudioapi::getActiveDocumentContext()$path))
at the beginning of your script. R will then set the working directory
to the folder that the script you are working with is located in.
R Projects
I do not recommend using setwd(). Instead, I would
suggest using R projects. When working in RStudio,
using R Projects is one of the best ways to keep your
work organized, portable, and efficient. An R Project
is essentially a self-contained workspace that helps manage files,
working directories, and settings automatically.
There are several reasons why I prefer R projects:
1️⃣ Automatic Working Directory Management
When you open an R Project, RStudio automatically
sets the working directory to the project’s folder. This means you don’t
have to use setwd() manually or worry about absolute file
paths.
Example:
- If your project folder is
C:/Users/YourName/Documents/MyProject, then any file in
this folder can be accessed with a relative path:
R
read.csv("data/my_data.csv") # No need for long paths!
2️⃣ Keeps Everything Organized
An R Project keeps all related files—scripts, datasets, plots, and outputs—in one place. This is especially useful when working on multiple projects, preventing files from getting mixed up.
A typical project folder might look like this:
MyProject/
│── data/ # Raw data files
│── scripts/ # R scripts
│── output/ # Processed results
│── MyProject.Rproj # The project fileThis structure helps keep your work clean and easy to navigate.
3️⃣ Easier Collaboration & Portability
If you share your project folder, others can open the
.Rproj file in RStudio and immediately start working—no
need to change file paths or set up the environment manually. This makes
R Projects ideal for:
✅ Teamwork
✅ Sharing with collaborators
✅ Reproducible research
How to Create an R Project
1️⃣ Open RStudio
2️⃣ Click File → New Project
3️⃣ Choose New Directory (or an existing folder)
4️⃣ Select New Project and give it a name
5️⃣ Click Create Project 🎉
Now, whenever you open the .Rproj file, RStudio will
automatically set everything up for you!
You can open a project using File > Open Project in
the top left of R Studio. You will then notice the projects name
appearing in the top right. Furthermore, the Files view in
the bottom right will automatically go to the destination of your
selected project.
Creating an R project
Let’s go through the steps of setting up an R project together.
First, decide if you want to use an existing folder or generate a brand
new folder for your R project. I would suggest using a folder that is
clean. For example, use the folder r_for_empra that you
created a couple lessons earlier!
To create a project, follow the steps outlined above. Make sure the creation of your project was successful and you can see the name of your project in the top right corner. If you are creating the project in a new folder, you will have to give it a sensible name!
Challenges
Challenge 1:
Make sure you have the project created. In the folder that the
project is created in, add the following sub-folders: data,
processed_data and archive. You can do this
either in the file explorer of your computer, or by using the bottom
right window of RStudio.
data will house all the raw data files that we are
working with in this tutorial. processed_data will be used
to save data that we processed in some way and want to reuse later on.
archive is a folder to store old scripts or data-sets that
you will no longer use. You can also delete those files, but I often
find it easier to just “shove them under the bed” so that I
could still use them later on.
Get an overview of the folders in your project by running
list.dirs(recursive = FALSE). This should return the
folders you just created and a folder where R-project information is
stored.
If you do not see the folders you just created, make sure you created
and opened the project and that the folders are in the same space as
your .Rproj project file.
Challenge 2:
Download the following data
and place it in the data folder. Make sure to give it an
appropriate name. The data contains information about the number of
times the character Roy Kent used the f-word in the show
Ted Lasso.
Challenge 3:
Test the data you just downloaded. Read it into R and give it an appropriate name.
Hint: You may need to research how to read .csv files
into R first.
R
roy_kent_data <- read.csv("data/roy_kent_f_count.csv")
Challenge 4:
Familiarize yourself with the data. What columns are there (use
colnames(data)). Compute the average amount of times Roy
Kent says “Fuck” in an episode. Compute the total amount of times he
said it throughout the series.
R
average_f_count <- mean(roy_kent_data$F_count_RK)
total_f_count <- sum(roy_kent_data$F_count_RK)
# Because, cum_rk_overall is a running (cumulative) total:
total_f_count <- max(roy_kent_data$cum_rk_overall)
- The working directory is where R looks for and
saves files.
- Absolute paths give full file locations; relative paths use the
working directory.
-
R Projects manage the working directory
automatically, keeping work organized.
- Using R Projects makes code portable, reproducible, and
easier to share.
- A structured project with separate folders for data and scripts improves workflow.
Content from Data Visualization (1)
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- How can you get an overview of data?
- How do you visualize distributions of data?
Objectives
- Learn how to read in
.csvfiles - Learn how to explore a dataset for the first time
- Learn how to visualize key statistics of data
Reading in data
You already had to read in data in the last lesson. Data comes in
many shapes and sizes. The format of data you had to read in the last
session is called csv, for comma-separated values.
This is one of the most common data formats. It gets its name because
all values are separated from each other using a comma “,”. Here in
Germany, csv files are sometimes separated by a semicolon
“;” instead. This can be adjusted in R-code by using
read.csv("path/to/file", sep = ";") and declaring the
semicolon as the separating variable.
Reading data requires specific functions
There are several other common formats including .xlsx
for excel files, .sav for SPSS files, .txt for
simple text files and .rdata for a special R-specific data
format. These files can not be read in by read.csv(), as
this is only for .csv files. Instead, use specific
functions for each format like readxl::read_excel(), or
haven::read_sav().
Let’s again read in the data about Roy Kent’s potty-mouth.
R
roy_kent_data <- read.csv("data/roy_kent_f_count.csv")

Getting a glimpse of the data
When starting out with some data you received, it is important to familiarize yourself with the basics of this data first. What columns does it have, what information is contained in each row? What do the columns mean?
One option to get a quick overview of the data is… just looking at
it! By clicking on the data in the Environment tab or typing
View(roy_kent_data) in the console, you can inspect the
data in R Studio. This will already show you how many observations (in
this case rows) and variables (columns) the data has. We can also
inspect the column names and see the first few entries. Look at the
data, what might each column represent?
Now, let’s go back to using programming to inspect data. We already
learned about the handy function glimpse(), for which we
need the dplyr package.
R
library(dplyr)
glimpse(roy_kent_data)
OUTPUT
Rows: 34
Columns: 16
$ Character <chr> "Roy Kent", "Roy Kent", "Roy Kent", "Roy Kent", "Roy…
$ Episode_order <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 1…
$ Season <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2…
$ Episode <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 1, 2, 3, 4, 5, 6, 7, …
$ Season_Episode <chr> "S1_e1", "S1_e2", "S1_e3", "S1_e4", "S1_e5", "S1_e6"…
$ F_count_RK <int> 2, 2, 7, 8, 4, 2, 5, 7, 14, 5, 11, 10, 2, 2, 23, 12,…
$ F_count_total <int> 13, 8, 13, 17, 13, 9, 15, 18, 22, 22, 16, 22, 8, 6, …
$ cum_rk_season <int> 2, 4, 11, 19, 23, 25, 30, 37, 51, 56, 11, 21, 23, 25…
$ cum_total_season <int> 13, 21, 34, 51, 64, 73, 88, 106, 128, 150, 16, 38, 4…
$ cum_rk_overall <int> 2, 4, 11, 19, 23, 25, 30, 37, 51, 56, 67, 77, 79, 81…
$ cum_total_overall <int> 13, 21, 34, 51, 64, 73, 88, 106, 128, 150, 166, 188,…
$ F_score <dbl> 0.1538462, 0.2500000, 0.5384615, 0.4705882, 0.307692…
$ F_perc <dbl> 15.4, 25.0, 53.8, 47.1, 30.8, 22.2, 33.3, 38.9, 63.6…
$ Dating_flag <chr> "No", "No", "No", "No", "No", "No", "No", "Yes", "Ye…
$ Coaching_flag <chr> "No", "No", "No", "No", "No", "No", "No", "No", "No"…
$ Imdb_rating <dbl> 7.8, 8.1, 8.5, 8.2, 8.9, 8.5, 9.0, 8.7, 8.6, 9.1, 7.…This will tell us the variable type of each column, too. We can see that the first column is a character column which just contains the name of the hero of this data Roy Kent. The second column is an integer (so whole numbers) vector with information about the episode number.
Next, let’s try to figure out what each of the column names mean and
what type of information is stored here. Some columns are
self-explanatory like Season or Episode.
Others might require a bit of background knowledge like
Imdb_rating, which requires you to know about the
film-rating platform “Imdb”.
Some columns are riddled with abbreviations that make their names
easier to write, but harder to understand. F_count_RK is
using RK as an abbreviation of Roy Kent and F as an abbreviation for…
something else. cum_rk_overall is a cumulative (running)
total of the number of times Roy used the F-word.
Yet other columns are really difficult to understand, because the
implication is not 100% clear. What does F_count_total
mean? The number of times “Fuck” was said in an episode? Seems like a
reasonable guess, but in order to be sure, it’s best if we had some
documentation of the data’s author’s intentions. This is often called a
codebook, where the person compiling data writes down
information about the meaning of column names, cleaning procedures and
other oddities of the data. When sharing your own data online, this is
essential in order to make your data useable for others.
There is no codebook included in this workshop, but you can find it online. The data is taken from an event called “Tidy-Tuesday”, a weekly event about cleaning and presenting interesting data using R. You can find information about the Roy Kent data here. Give it a read in order to make sure you know what all the columns are about.
Inspecting important values
Now that we understand the structure of the data and the names of the
columns, we can start getting a look at the data itself. A good way to
get an overview of data with loads of numeric variables is
psych::describe(). This outputs you some basic descriptive
statistics of the data.
R
psych::describe(roy_kent_data) # describe() function from psych package
OUTPUT
vars n mean sd median trimmed mad min max
Character* 1 34 1.00 0.00 1.00 1.00 0.00 1.00 1.00
Episode_order 2 34 17.50 9.96 17.50 17.50 12.60 1.00 34.00
Season 3 34 2.06 0.81 2.00 2.07 1.48 1.00 3.00
Episode 4 34 6.21 3.37 6.00 6.18 4.45 1.00 12.00
Season_Episode* 5 34 17.50 9.96 17.50 17.50 12.60 1.00 34.00
F_count_RK 6 34 8.82 5.63 7.50 8.25 5.19 2.00 23.00
F_count_total 7 34 21.82 11.53 18.00 20.86 8.15 6.00 46.00
cum_rk_season 8 34 53.79 37.29 50.00 51.36 40.77 2.00 138.00
cum_total_season 9 34 126.32 93.71 110.50 115.79 88.96 13.00 377.00
cum_rk_overall 10 34 130.74 92.69 124.50 127.25 118.61 2.00 300.00
cum_total_overall 11 34 308.09 215.14 275.50 296.61 246.11 13.00 742.00
F_score 12 34 0.40 0.15 0.36 0.38 0.15 0.15 0.72
F_perc 13 34 39.57 15.02 35.70 38.49 15.20 15.40 71.90
Dating_flag* 14 34 1.44 0.50 1.00 1.43 0.00 1.00 2.00
Coaching_flag* 15 34 1.59 0.50 2.00 1.61 0.00 1.00 2.00
Imdb_rating 16 34 8.33 0.66 8.50 8.39 0.59 6.60 9.40
range skew kurtosis se
Character* 0.00 NaN NaN 0.00
Episode_order 33.00 0.00 -1.31 1.71
Season 2.00 -0.10 -1.53 0.14
Episode 11.00 0.06 -1.26 0.58
Season_Episode* 33.00 0.00 -1.31 1.71
F_count_RK 21.00 0.79 0.05 0.97
F_count_total 40.00 0.78 -0.58 1.98
cum_rk_season 136.00 0.48 -0.82 6.39
cum_total_season 364.00 0.93 0.16 16.07
cum_rk_overall 298.00 0.28 -1.27 15.90
cum_total_overall 729.00 0.42 -1.06 36.90
F_score 0.56 0.60 -0.74 0.03
F_perc 56.50 0.60 -0.74 2.58
Dating_flag* 1.00 0.23 -2.01 0.09
Coaching_flag* 1.00 -0.34 -1.94 0.09
Imdb_rating 2.80 -0.81 0.31 0.11In my opinion, the most important information in this table are the mean value of a column and the maximum and minimum value. The mean value reveals some important information about the average value of a given variable, duh… And the minimum and maximum value can reveal issues in the data. Is the minimum age -2? Or the maximum IQ 1076? Something might be wrong in this data, and looking at the minimum and maximum values might already give you a clue. In this data, none of the maxima or minima seem to be a problem, as they are all in reasonable ranges.
Let’s start visualizing some basic information. I have already proclaimed my love for histograms as a quick and dirty tool to get an overview and the simplicity of its code is a reason why!
R
hist(roy_kent_data$F_count_RK) # Recall that the $ operator gets you a variable from a dataframe
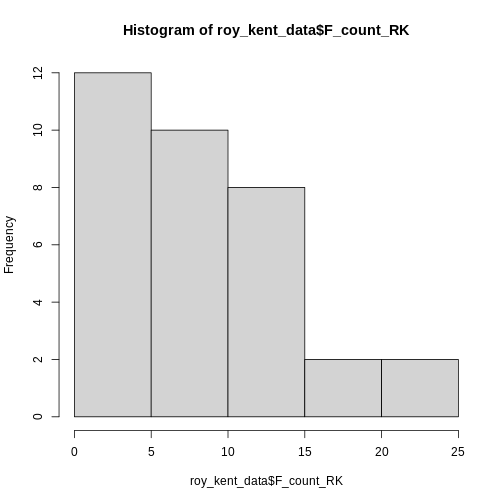
Only 5 bars seem to be to few to visualize the distribution adequately, so lets try 10 instead.
R
hist(roy_kent_data$F_count_RK, breaks = 10)
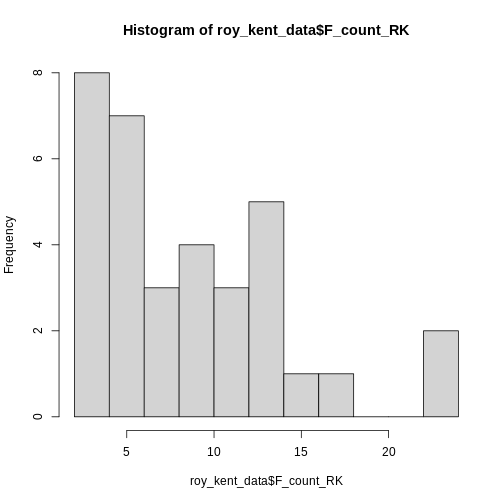 This already provides us some more useful information. Roy seems to
swear between 0 and 12 times most often and swears a lot in two
episodes.
This already provides us some more useful information. Roy seems to
swear between 0 and 12 times most often and swears a lot in two
episodes.
Let’s try increasing the number of breaks to 20.
R
hist(roy_kent_data$F_count_RK, breaks = 20)
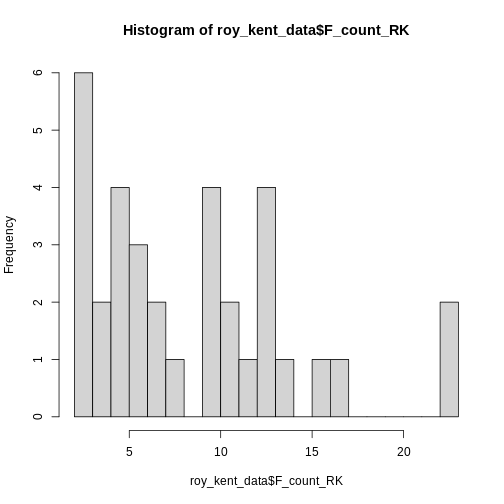
Here, an interesting pattern seems to emerge. Three different
clusters of swear counts show up, one where Roy swears between 0 - 7
times, presumably because he has little screen-time in the episode. One
cluster where Roy swears between 9-17 times, which more closely
resembles his average swearing amount. And finally two episodes where
Roy flies of the handle and swears a whopping
max(roy_kent_data$F_count_RK) = 23 times!
In order to figure out what makes Roy swear so much, let’s plot the number of times Roy swears by episode!
ggplot2
For most of our plotting needs (except the beautiful
hist()), we will make use of the package
ggplot2 which is part of the larger collection of packages
tidyverse and provides some really useful plotting
functions. Most importantly, ggplot provides a useful grammar
of plotting functions that always follow the same format.
You start out with the function ggplot() and give it our
data, simple enough. Here, we start working with the entire dataframe,
not just vectors. We can simply pass it into ggplot() and
later on declare which variables we want to use.
R
ggplot(data = roy_kent_data)
ERROR
Error in ggplot(data = roy_kent_data): could not find function "ggplot"Oops, R tells us that it cannot find this function. Most of the time, this is because you forgot to load the package beforehand. Let’s try again:
R
library(ggplot2)
ggplot(data = roy_kent_data)

Now it works, but not really. For now, this code only tells the function which data we are using, not what to do with it.
In order to actually see a plot, we need to provide information about
the visual properties, the aesthetics that the plot should
have. The mapping argument tells the function how the data
relates to the visual properties of the plot. We define this mapping
using the function aes(). Here, we declare the columns that
we want on the x-axis and y-axis.
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK)
)
While we cannot see anything useful yet, we already notice some changes. The plot now correctly shows the names of the variables on the axes. Also, the ranges of numbers printed on the axes matches the minimum and maximum values.
Now that we provided the data and the mapping of variables to the
plot, we can start building our proper visualization. This is done
on top of the basics declared in ggplot(). This
technique is called layering and it allows you to combine
multiple new changes to a plot easily using a simple +. To
add a plot showing the number of Fucks given by Roy during an episode,
we can use geom_point().
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK)
) +
geom_point()
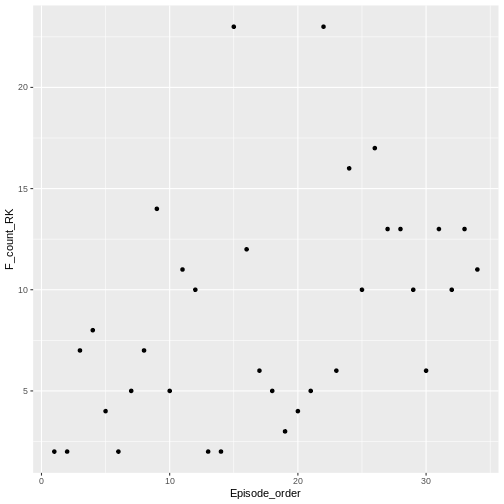
geom_point() adds a simple scatter-plot element to the
graph.
We can change the labels to make the graph more polished using
labs().
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK)
) +
geom_point()+
labs(
title = "# of F*cks said by Roy Kent",
x = "Episode Number",
y = "# of F*cks"
)
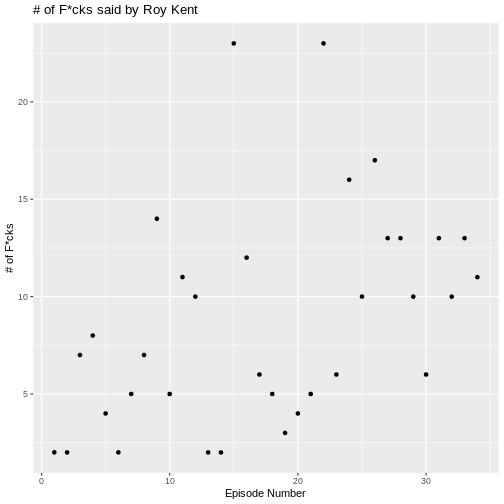
There seems to be a small trend showing that Roy swears more later in
the series. We can visualize this trend by adding another layered visual
geom_smooth().
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK)
) +
geom_point()+
labs(
title = "# of F*cks said by Roy Kent",
x = "Episode Number",
y = "# of F*cks"
)+
geom_smooth(
method = "lm", # this forces the trend-line to be linear
se = FALSE
)
OUTPUT
`geom_smooth()` using formula = 'y ~ x'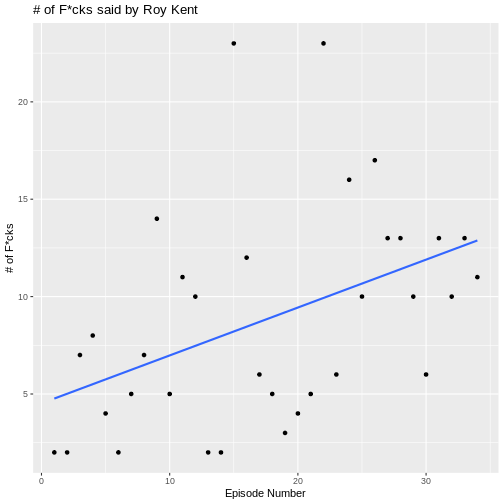
What might be causing this trend? Well, one possible candidate is
that Roy goes from being a player on the team to being a coach on the
team. This might cause him to yell at more people than usual. The
variable that contains this information is Coaching_flag.
Let’s learn something about it:
R
table(roy_kent_data$Coaching_flag)
OUTPUT
No Yes
14 20 It seems to have two values “Yes” and “No”, each being represented in the data more than 10 times.
We can also add this to our visualization from earlier, coloring in
the dots by whether Roy was coaching or not. In order to do this, we
need to add the color variable in the aesthetics mapping in
the ggplot() function.
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK, color = Coaching_flag)
) +
geom_point()+
labs(
title = "# of F*cks said by Roy Kent",
x = "Episode Number",
y = "# of F*cks"
)+
geom_smooth(
method = "lm", # this forces the trend-line to be linear
se = FALSE
)
OUTPUT
`geom_smooth()` using formula = 'y ~ x'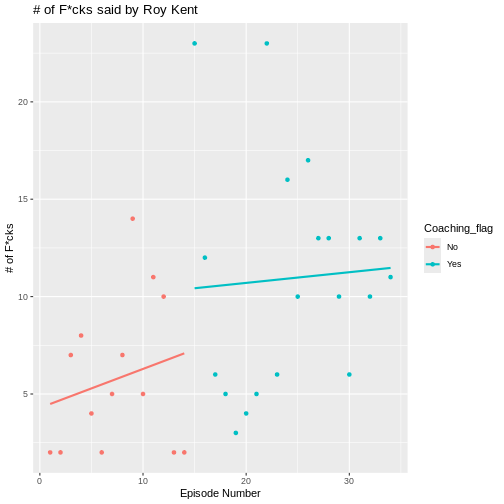
Now, a clearer picture emerges. Roy starts of pretty slow as a player, but then begins to swear a lot in the episodes that he is coaching in.
Making plots prettier using themes
The basic plots already look okay and are enough for just finding something out about data. But you can make them even more enticing by using a theme. Themes in ggplot are collections of visual settings that control the background of the plot, the text size of the axis and some other things.
I often use theme_classic() or
theme_minimal(), but you can try out different themes or
even write your own!
R
ggplot(
data = roy_kent_data,
mapping = aes(x = Episode_order, y = F_count_RK, color = Coaching_flag)
) +
geom_point()+
labs(
title = "# of F*cks said by Roy Kent",
x = "Episode Number",
y = "# of F*cks"
)+
geom_smooth(
method = "lm", # this forces the trend-line to be linear
se = FALSE
)+
theme_minimal()
OUTPUT
`geom_smooth()` using formula = 'y ~ x'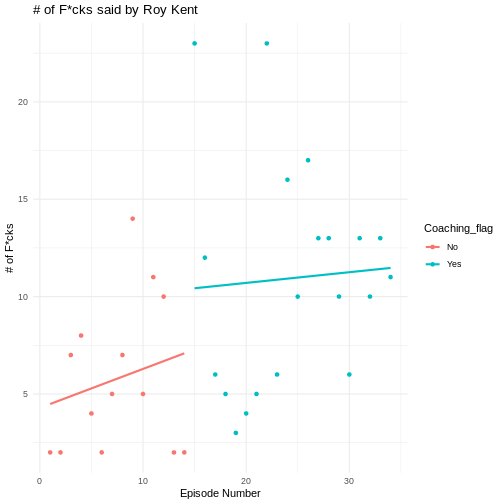
Challenges
Challenge 1
Figure out what information is stored in the column
F_count_total. Compute the descriptive information (mean,
standard deviation, minimum and maximum) for this variable.
Challenge 2
Compare the descriptive information to those for
F_count_RK. Which one is bigger, why?
Compute the difference between the two variables. What does it
represent. Compare the difference to F_count_RK. Which has
the higher average value?
R
f_count_others <- roy_kent_data$F_count_total - roy_kent_data$F_count_RK
mean(f_count_others)
OUTPUT
[1] 13R
mean(roy_kent_data$F_count_RK)
OUTPUT
[1] 8.823529Challenge 3
Plot a histogram of F_count_total. Try different values
of breaks, which seems most interesting to you?
Challenge 4
Visualize the relationship between the amount of time Roy Kent said
“Fuck” and the Imdb rating of that episode using
geom_point().
R
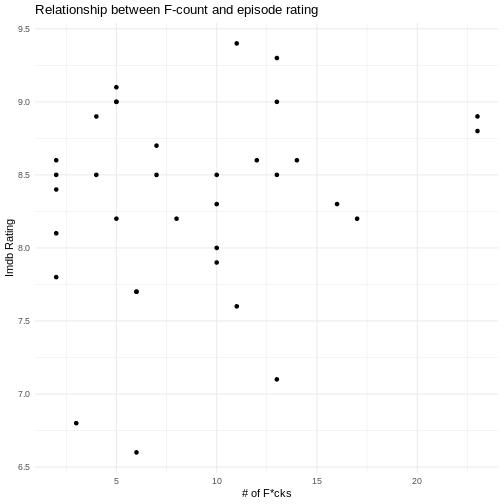
ggplot(
data = roy_kent_data,
mapping = aes(x = F_count_RK, y = Imdb_rating)
)+
geom_point()+
labs(
title = "Relationship between F-count and episode rating",
x = "# of F*cks",
y = "Imdb Rating"
)+
theme_minimal()
Challenge 5
Be creative! What else can you notice in the data? Try to make an interesting plot.
- Get an overview of the data by inspecting it or using
glimpse()/describe() - Consult a codebook for more in-depth descriptions of the variables in the data
- Visualize the distribution of a variable using
hist() - Use
ggplot(data = data, mapping = aes())to provide the data and mapping to the plots - Add visualization steps like
geom_point()orgeom_smooth()using+
Content from Data Visualization (2)
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- How can you visualize categorical data?
- How to visualize the relationship of categorical and numerical data?
Objectives
- Get to know the DASS data from Kaggle
- Explain how to plot distribution of numerical data using
geom_histogram()andgeom_density() - Demonstrate how to group distributions by categorical variables
A new set of data
In this section, we will work with a new set of data. Download this
data here and place it in your
data folder. There is also a codebook available to download
here.
The data originates from an online version of the Depression Anxiety Stress Scales (DASS), collected between 2017-2019 and retrieved from Kaggle. Participants took the test to receive personalized results and could opt into a research survey. The dataset includes responses to 42 DASS items (rated on a 4-point scale), response times, and question positions. Additional data includes:
-
Demographics (age, gender, education, native
language, handedness, religion, sexual orientation, race, marital
status, family size, major, country).
-
Big Five Personality Traits (via the Ten Item
Personality Inventory - TIPI).
- Vocabulary Check (includes fake words for validity checks).
Let’s read in the data first. In my case, it is placed inside a
subfolder of data/ called kaggle_dass/. You
may need to adjust the path on your end.
R
dass_data <- read.csv("data/kaggle_dass/data.csv")
As you might already see, this data is quite large. In the Environment tab we can see that it contains almost 40.000 entries and 172 variables. For now, we will be working with some of the demographic statistics. We will delve into the actual score in a later episode. You can find information on this data in the codebook provided here.
Read this codebook in order to familiarize yourself with the data.
Then inspect it visually by clicking on the frame or running
View(dass_data). What do you notice?
First, let’s start getting an overview of the demographics of
test-takers. What is their gender, age and education? Compute
descriptive statistics for age and inspect gender and education using
table()
R
table(dass_data$gender)
OUTPUT
0 1 2 3
67 8789 30367 552 R
mean(dass_data$age)
OUTPUT
[1] 23.61217R
sd(dass_data$age)
OUTPUT
[1] 21.58172R
range(dass_data$age) # This outputs min() and max() at the same time
OUTPUT
[1] 13 1998R
table(dass_data$education)
OUTPUT
0 1 2 3 4
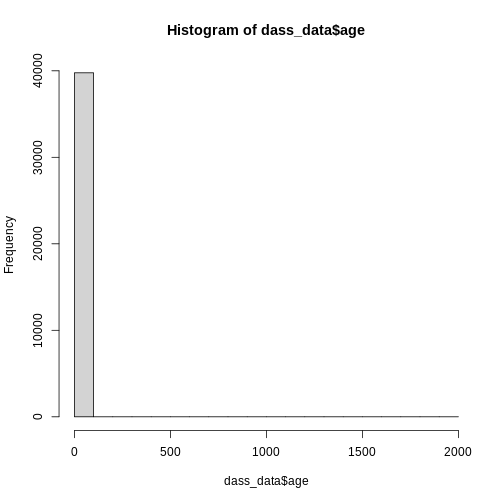
515 4066 15066 15120 5008 The maximum value of age seems a bit implausible, unless Elrond has decided to take the DASS after Galadriel started flirting with Sauron (sorry, Annatar) again.
Histograms
We can get an overview of age by using our trust
hist():
R
hist(dass_data$age)
However, this graph is heavily skewed by the outliers in age. We can
address this issue easily by converting to a ggplot
histogram. The xlim() layer can restrict the values that
are displayed in the plot and gives us a warning about how many values
were discarded.
R
library(ggplot2) # Don't forget to load the package
ggplot(
data = dass_data,
mapping = aes(x = age)
)+
geom_histogram()+
xlim(0, 100)
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_bin()`).WARNING
Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).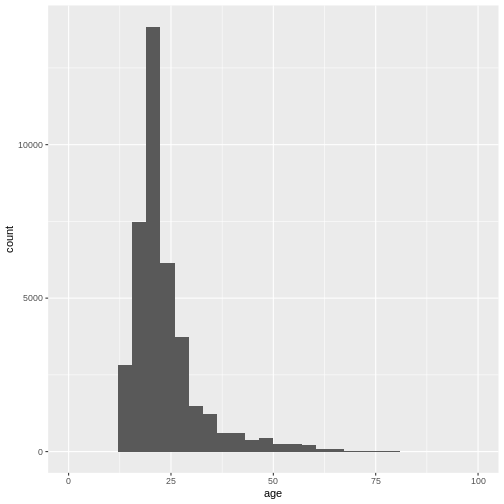
Again, the number of bins might be a bit small:
R
ggplot(
data = dass_data,
mapping = aes(x = age)
)+
geom_histogram(bins = 100)+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_bin()`).WARNING
Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).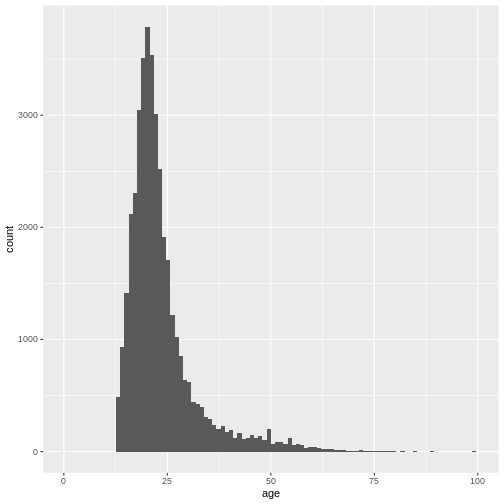 We can also adjust the size of the bins, not the number of bins by using
the
We can also adjust the size of the bins, not the number of bins by using
the binwidth argument.
R
ggplot(
data = dass_data,
mapping = aes(x = age)
)+
geom_histogram(binwidth = 1)+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_bin()`).WARNING
Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).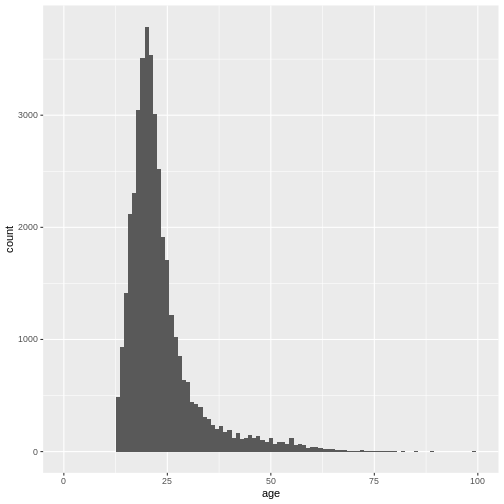
This gives us a nice overview of the distribution of each possible distinct age. It seems to peak around 23 and then slowly decline in frequency. This is in line with what you would expect from data from a free online personality questionnaire.
Density plots
Often, what interests us is not the number of occurrences for a given
value, but rather which values are common and which values are uncommon.
By dividing the number of occurrences for a given value by the total
number of observation, we can obtain a density-plot. In
ggplot you achieve this by using
geom_density() instead of
geom_histogram().
R
ggplot(
data = dass_data,
mapping = aes(x = age)
)+
geom_density()+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_density()`).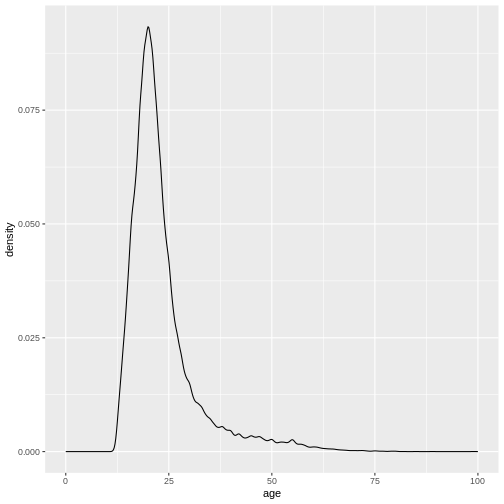
This describes the overall age distribution, but we can also look at the age distribution by education status. What would you expect?
Again, we do this by using the color argument in
aes(), as I want different colored density plots for each
education level.
R
ggplot(
data = dass_data,
mapping = aes(x = age, color = education)
)+
geom_density()+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_density()`).WARNING
Warning: The following aesthetics were dropped during statistical transformation:
colour.
ℹ This can happen when ggplot fails to infer the correct grouping structure in
the data.
ℹ Did you forget to specify a `group` aesthetic or to convert a numerical
variable into a factor?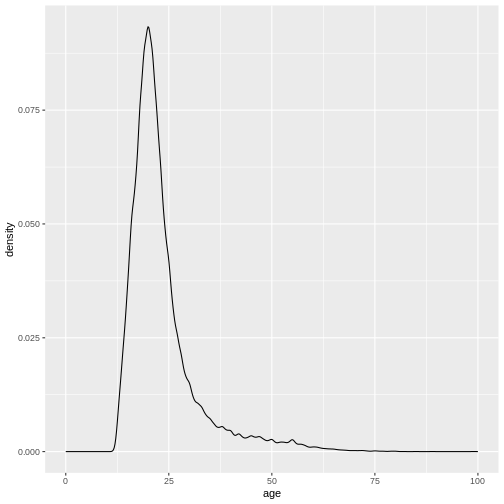
It seems like this didn’t work. And the warning message tells us why.
We forgot to specify a group variable that tells the
plotting function that we not only want different colors, we also want
different lines, grouped by education status:
R
ggplot(
data = dass_data,
mapping = aes(x = age, color = education, group = education)
)+
geom_density()+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_density()`).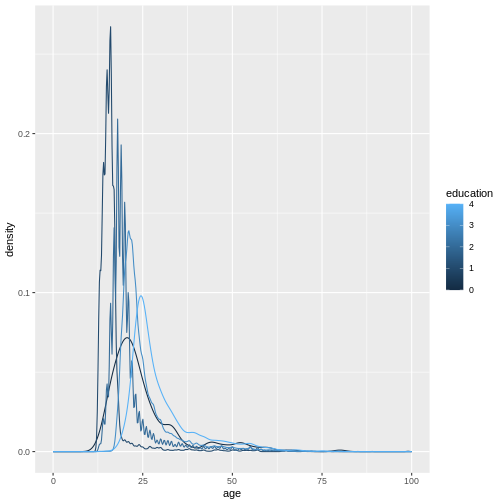
Because education is a numerical variable, ggplot uses a
sliding color scale to color its values. I think it looks a little more
beautiful if we declare that despite having numbers as entries,
education is not strictly numerical. The different numbers
represent distinct levels of educational attainment. We can encode this
using factor() to convert education into a
variable-type called factor that has this property.
R
dass_data$education <- factor(dass_data$education)
ggplot(
data = dass_data,
mapping = aes(x = age, color = education, group = education)
)+
geom_density()+
xlim(0, 100)
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_density()`).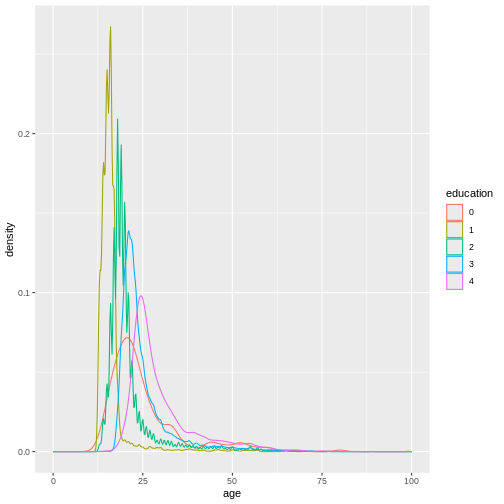
We can make this graph look a little bit better by filling in the
area under the curves with the color aswell and making them slightly
transparent. This can be done using the fill argument and
the alpha argument. Let’s also give the graph a proper
title and labels.
R
ggplot(
data = dass_data,
mapping = aes(x = age, color = education, group = education, fill = education)
)+
geom_density(alpha = 0.5)+
xlim(0, 100)+
labs(
title = "Age distribution by education status",
x = "Age",
y = "Density"
) +
theme_minimal()
WARNING
Warning: Removed 7 rows containing non-finite outside the scale range
(`stat_density()`).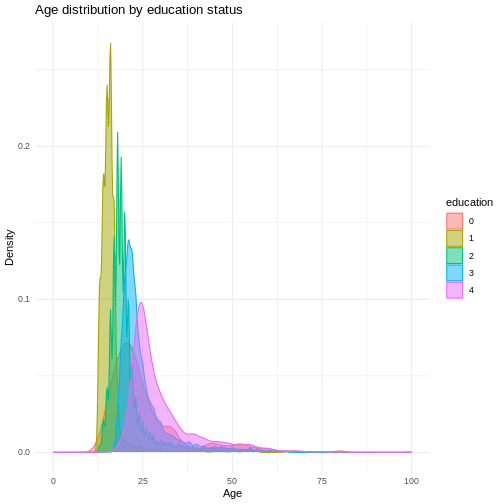
In this graph, we can see exactly what we would expect. As the highest achieved education increases, the age of test-takers generally gets larger as-well. There just are very little people under 20 with a PhD.
Bar Charts
As a last step of exploration, let’s visualize the number of people that achieved a certain educational status. A bar chart is perfect for this:
R
ggplot(
data = dass_data,
mapping = aes(x = education)
)+
geom_bar()
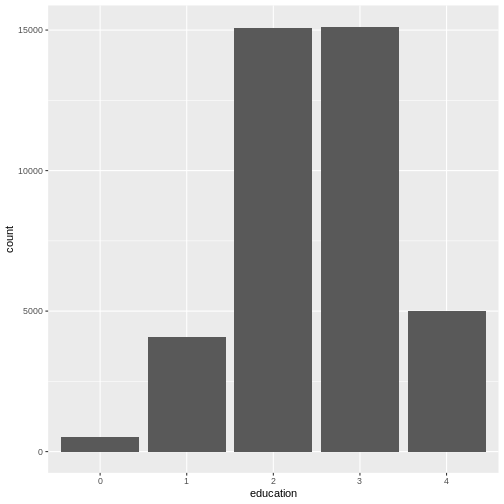
This visualizes the same numbers as
table(dass_data$education) and sometimes, you may not need
to create a plot if the numbers already tell you everything you
need to know. However, remember that often you need to communicate your
results to other people. And as they say “A picture says more than 1000
words”.
We can also include gender as a variable here, in order to compare educational attainment between available genders. Let’s plot gender on the x-axis and color the different education levels.
R
ggplot(
data = dass_data,
mapping = aes(x = gender, fill = education)
)+
geom_bar()
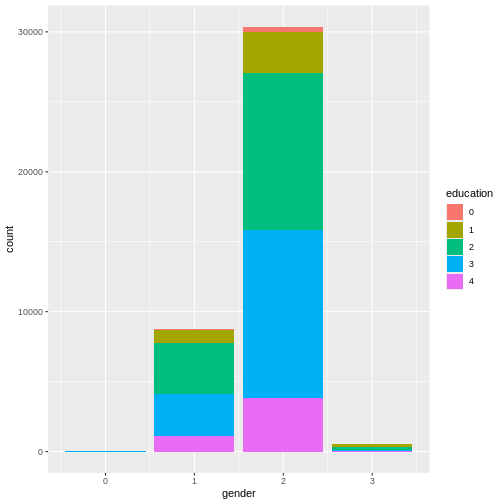
This graph is very good at showing one thing: there are more females
in the data than males. However, this is not the comparison we are
interested in. We can plot relative frequencies of education by gender
by using the argument position = "fill".
R
ggplot(
data = dass_data,
mapping = aes(x = gender, fill = education)
)+
geom_bar(position = "fill")
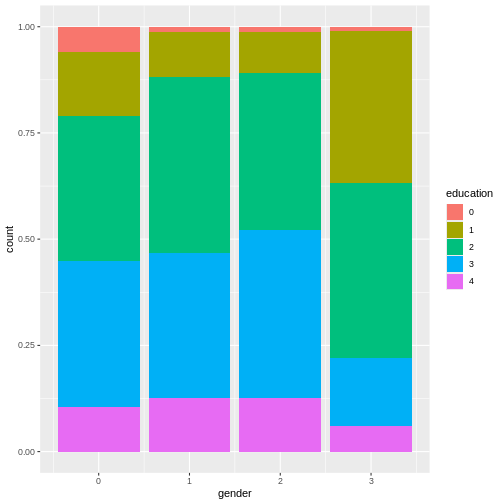
This plot shows that education levels don’t differ dramatically between males and females in our sample, with females holding a university degree more often than males.
Common Problems
This section is lifted from R for Data Science (2e).
As you start to run R code, you’re likely to run into problems. Don’t worry — it happens to everyone. We have all been writing R code for years, but every day we still write code that doesn’t work on the first try!
Start by carefully comparing the code that you’re running to the code
in the book. R is extremely picky, and a misplaced character can make
all the difference. Make sure that every ( is matched with
a ) and every " is paired with another
". Sometimes you’ll run the code and nothing happens. Check
the left-hand of your console: if it’s a +, it means that R
doesn’t think you’ve typed a complete expression and it’s waiting for
you to finish it. In this case, it’s usually easy to start from scratch
again by pressing ESCAPE to abort processing the current command.
One common problem when creating ggplot2 graphics is to put the + in the wrong place: it has to come at the end of the line, not the start. In other words, make sure you haven’t accidentally written code like this:
R
ggplot(data = mpg)
+ geom_point(mapping = aes(x = displ, y = hwy))
If you’re still stuck, try the help. You can get help about any R
function by running ?function_name in the console, or
highlighting the function name and pressing F1 in RStudio. Don’t worry
if the help doesn’t seem that helpful - instead skip down to the
examples and look for code that matches what you’re trying to do.
If that doesn’t help, carefully read the error message. Sometimes the answer will be buried there! But when you’re new to R, even if the answer is in the error message, you might not yet know how to understand it. Another great tool is Google: try googling the error message, as it’s likely someone else has had the same problem, and has gotten help online.
Recap - Knowing your data!
It’s important to understand your data, its sources and quirks before you start working with it! This is why the last few episodes focused so much on descriptive statistics and plots. Get to know the structure of your data, figure out where it came from and what type of people constitute the sample. Monitor your key variables for implausible or impossible values. Statistical outliers we can define later, but make sure to identify issues with impossible values early on!
Challenges
Challenge 1
Review what we learned about the DASS data so far. What are the key demographic indicators? Is there anything else important to note?
Challenge 3
Provide descriptive statistics of the time it took participants to complete the DASS-part of the survey. What variable is this stored in? What is the average time? Are there any outliers? What is the average time without outliers?
Challenge 4
Plot a distribution of the elapsed test time. What might be a sensible cutoff for outliers?
Plot a distribution of elapsed test time by whether English was the native language. What do you expect? What can you see? What are your thoughts on the distribution?
R
dass_data$engnat <- factor(dass_data$engnat)
ggplot(
data = dass_data,
mapping = aes(
x = testelapse, fill = engnat, group = engnat
)
)+
geom_density(alpha = 0.5)+
xlim(0, 3000)+
labs(
title = "Distribution of test completion times",
subtitle = "By Mother Tongue",
x = "Test completion time (s)",
y = "Density"
)+
theme_minimal()
WARNING
Warning: Removed 399 rows containing non-finite outside the scale range
(`stat_density()`).
Challenge 5
Learn something about the handedness of test-takers. Get the number
of test-takers with each handedness using table and
visualize it using geom_bar().
Plot the handedness of test-takers by educational status. Do you see any differences?
R
ggplot(
data = dass_data,
mapping = aes(
x = hand,
fill = education,
group = education
)
)+
geom_bar(position = "fill")+
labs(
title = "Educational attainment by handedness",
x = "Handedness (0 = NA, 1 = Right, 2 = Left, 3 = Both)",
y = "Proportion"
)+
theme_minimal()
- Get to know new data by inspecting it and computing key descriptive statistics
- Visualize distributions of key variables in order to learn about factors that impact them
- Visualize distribution of a numeric and a categorical variable using
geom_density() - Visualize distribution of two categorial variables using
geom_bar()
Content from Filtering data
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- How do you filter invalid data?
- How do you chain commands using the pipe
%>%? - How can I select only some columns of my data?
Objectives
- Explain how to filter rows using
filter() - Demonstrate how to select columns using
select() - Explain how to use the pipe
%>%to facilitate clean code
Filtering
In the earlier episodes, we have already seen some impossible values
in age and testelapse. In our analyses, we
probably want to exclude those values, or at the very least investigate
what is wrong. In order to do that, we need to filter our data
- to make sure it only has those observations that we actually want. The
dplyr package provides a useful function for this:
filter().
Let’s start by again loading in our data, and also loading the
package dplyr.
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
The filter function works with two key inputs, the data
and the arguments that you want to filter by. The data is always the
first argument in the filter function. The following arguments are the
filter conditions. For example, in order to only include people that are
20 years old, we can run the following code:
R
twenty_year_olds <- filter(dass_data, age == 20)
# Lets make sure this worked
table(twenty_year_olds$age)
OUTPUT
20
3789 R
# range(twenty_year_olds$age) # OR THIS
# unique(twenty_year_olds$age) # OR THIS
# OR other things that come to mind can check this
Now, in order to only include participants aged between 0 and 100,
which seems like a reasonable range, we can condition on
age >= 0 & age <= 100.
R
valid_age_dass <- filter(dass_data, age >= 0 & age <= 100)
Let’s check if this worked using a histogram.
R
hist(valid_age_dass$age, breaks = 50)
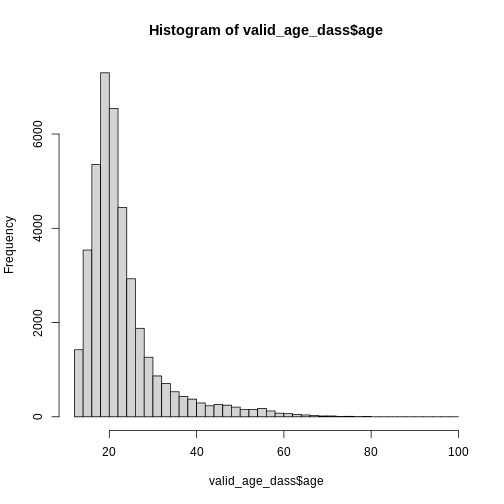
Looks good! Now it is always a good idea to inspect the values you removed from the data, in order to make sure that you did not remove something you intended to keep, and to learn something about the data you removed.
Let’s first check how many rows of data we actually removed:
R
n_rows_original <- nrow(dass_data)
n_rows_valid_age <- nrow(valid_age_dass)
n_rows_removed <- n_rows_original - n_rows_valid_age
n_rows_removed
OUTPUT
[1] 7This seems okay! Only 7 removed cases out of almost 40000 entries is negligible. Nonentheless, let’s investigate this further.
R
impossible_age_data <- filter(dass_data, age < 0 | age > 100)
Now we have a dataset that contains only the few cases with invalid ages. We can just look at the ages first:
R
impossible_age_data$age
OUTPUT
[1] 223 1996 117 1998 115 1993 1991Now, let’s try investigating the a little deeper. Do the people with invalid entries in age have valid entries in the other columns? Try to figure this out by visually inspecting the data. Where are they from? Do the responses to other questions seem odd?
This is possible, but somewhat hard to do for data with 170 columns. Looking through all of them might make you a bit tired after the third investigation of invalid data. What we really need is to look at only a few columns that interest us.
Selecting columns
This is where the function select() comes in. It enables
us to only retain some columns from a dataset for further analysis. You
just give it the data as the first argument and then list all of the
columns you want to keep.
R
inspection_data_impossible_age <- select(
impossible_age_data,
age, country, engnat, testelapse
)
inspection_data_impossible_age
OUTPUT
age country engnat testelapse
1 223 MY 2 456
2 1996 MY 2 259
3 117 US 1 114
4 1998 MY 2 196
5 115 AR 2 238
6 1993 MY 2 232
7 1991 MY 2 240Here, we can see that our original data is reduced to the four
columns we specified above
age, country, engnat, testelapse. Do any of them look
strange? What happened in the age column?
To me, none of the other entries seem strange, the time it took them to complete the survey is plausible, they don’t show a clear trend of being from the same country. It seems that some people mistook the age question and entered their year of birth instead. Later on, we can try to fix this.
But before we do that, there are some more things about
select() that we can learn. It is one of the most useful
functions in my daily life, as it allows me to keep only those columns
that I am actually interested in. And it has many more features than
just listing the columns!
For example, note that the last few columns of the data are all the
demographic information. This is starting with education
and ending with major.
To select everything from education to
major:
R
# colnames() gives us the names of the columns only
colnames(select(valid_age_dass, education:major))
OUTPUT
[1] "education" "urban" "gender"
[4] "engnat" "age" "screensize"
[7] "uniquenetworklocation" "hand" "religion"
[10] "orientation" "race" "voted"
[13] "married" "familysize" "major" We can also select all character columns using
where():
R
colnames(select(valid_age_dass, where(is.character)))
OUTPUT
[1] "country" "major" Or we can select all columns that start with the letter “Q”:
R
colnames(select(valid_age_dass, starts_with("Q")))
OUTPUT
[1] "Q1A" "Q1I" "Q1E" "Q2A" "Q2I" "Q2E" "Q3A" "Q3I" "Q3E" "Q4A"
[11] "Q4I" "Q4E" "Q5A" "Q5I" "Q5E" "Q6A" "Q6I" "Q6E" "Q7A" "Q7I"
[21] "Q7E" "Q8A" "Q8I" "Q8E" "Q9A" "Q9I" "Q9E" "Q10A" "Q10I" "Q10E"
[31] "Q11A" "Q11I" "Q11E" "Q12A" "Q12I" "Q12E" "Q13A" "Q13I" "Q13E" "Q14A"
[41] "Q14I" "Q14E" "Q15A" "Q15I" "Q15E" "Q16A" "Q16I" "Q16E" "Q17A" "Q17I"
[51] "Q17E" "Q18A" "Q18I" "Q18E" "Q19A" "Q19I" "Q19E" "Q20A" "Q20I" "Q20E"
[61] "Q21A" "Q21I" "Q21E" "Q22A" "Q22I" "Q22E" "Q23A" "Q23I" "Q23E" "Q24A"
[71] "Q24I" "Q24E" "Q25A" "Q25I" "Q25E" "Q26A" "Q26I" "Q26E" "Q27A" "Q27I"
[81] "Q27E" "Q28A" "Q28I" "Q28E" "Q29A" "Q29I" "Q29E" "Q30A" "Q30I" "Q30E"
[91] "Q31A" "Q31I" "Q31E" "Q32A" "Q32I" "Q32E" "Q33A" "Q33I" "Q33E" "Q34A"
[101] "Q34I" "Q34E" "Q35A" "Q35I" "Q35E" "Q36A" "Q36I" "Q36E" "Q37A" "Q37I"
[111] "Q37E" "Q38A" "Q38I" "Q38E" "Q39A" "Q39I" "Q39E" "Q40A" "Q40I" "Q40E"
[121] "Q41A" "Q41I" "Q41E" "Q42A" "Q42I" "Q42E"There are a number of helper functions you can use within
select():
-
starts_with("abc"): matches names that begin with “abc”. -
ends_with("xyz"): matches names that end with “xyz”. -
contains("ijk"): matches names that contain “ijk”. -
num_range("x", 1:3): matchesx1,x2andx3.
You can also remove columns using select by simply negating the
argument using !:
R
colnames(select(valid_age_dass, !starts_with("Q")))
OUTPUT
[1] "country" "source" "introelapse"
[4] "testelapse" "surveyelapse" "TIPI1"
[7] "TIPI2" "TIPI3" "TIPI4"
[10] "TIPI5" "TIPI6" "TIPI7"
[13] "TIPI8" "TIPI9" "TIPI10"
[16] "VCL1" "VCL2" "VCL3"
[19] "VCL4" "VCL5" "VCL6"
[22] "VCL7" "VCL8" "VCL9"
[25] "VCL10" "VCL11" "VCL12"
[28] "VCL13" "VCL14" "VCL15"
[31] "VCL16" "education" "urban"
[34] "gender" "engnat" "age"
[37] "screensize" "uniquenetworklocation" "hand"
[40] "religion" "orientation" "race"
[43] "voted" "married" "familysize"
[46] "major" Now we will only get those columns that don’t start with “Q”.
Selecting a column twice
You can also use select to reorder the columns in your dataset. For example, notice that the following two bits of code return differently order data:
R
# head() gives us the first 10 values only
head(select(valid_age_dass, age, country, testelapse), 10)
OUTPUT
age country testelapse
1 16 IN 167
2 16 US 193
3 17 PL 271
4 13 US 261
5 19 MY 164
6 20 US 349
7 17 MX 45459
8 29 GB 232
9 16 US 195
10 18 DE 120R
head(select(valid_age_dass, testelapse, age, country), 10)
OUTPUT
testelapse age country
1 167 16 IN
2 193 16 US
3 271 17 PL
4 261 13 US
5 164 19 MY
6 349 20 US
7 45459 17 MX
8 232 29 GB
9 195 16 US
10 120 18 DEUsing the function everything(), you can select all
columns.
What happens, when you select a column twice? Try running the following examples, what do you notice about the position of columns?
R
select(valid_age_dass, age, country, age)
select(valid_age_dass, age, country, everything())
Using the pipe %>%
Let’s retrace some of the steps that we took. We had the original
data dass_data and then filtered out those rows with
invalid entries in age. Then, we tried selecting only a couple of
columns in order to make the data more manageable. In code:
R
impossible_age_data <- filter(dass_data, age < 0 | age > 100)
inspection_data_impossible_age <- select(
impossible_age_data,
age, country, engnat, testelapse
)
Notice how the first argument for both the filter() and
the select() function is always the data. This is also true
for the ggplot() function. And it’s no coincidence! In R,
there exists a special symbol, called the pipe %>%, that
has the following property: It takes the output of the previous function
and uses it as the first input in the following function.
This can make our example considerably easier to type:
R
dass_data %>%
filter(age < 0 | age > 100) %>%
select(age, country, engnat, testelapse)
OUTPUT
age country engnat testelapse
1 223 MY 2 456
2 1996 MY 2 259
3 117 US 1 114
4 1998 MY 2 196
5 115 AR 2 238
6 1993 MY 2 232
7 1991 MY 2 240Notice how the pipe is always written at the end of a line. This
makes it easier to understand and to read. As we go to the next line,
whatever is outputted is used as input in the following line. So the
original data dass_data is used as the first input in the
filter function, and the filtered data is used as the first input in the
select function.
To use the pipe, you can either type it out yourself or use the
Keyboard-Shortcut Ctrl + Shift + M.
Plotting using the pipe
We have seen that the pipe %>% allows us to chain
multiple functions together, making our code cleaner and easier to read.
This can be particularly useful when working with ggplot2,
where we often build plots step by step. Since ggplot()
takes the data as its first argument, we can use the pipe to make our
code more readable.
For example, let’s say we want to create a histogram of valid ages in our dataset. Instead of writing:
R
library(ggplot2)
ggplot(valid_age_dass, aes(x = age)) +
geom_histogram(bins = 50)
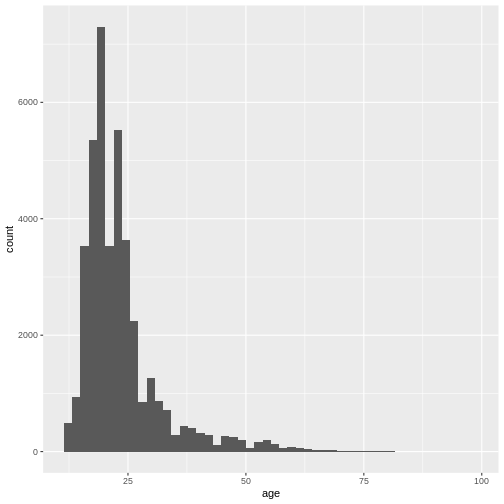
We can write:
R
valid_age_dass %>%
ggplot(aes(x = age)) +
geom_histogram(bins = 50)
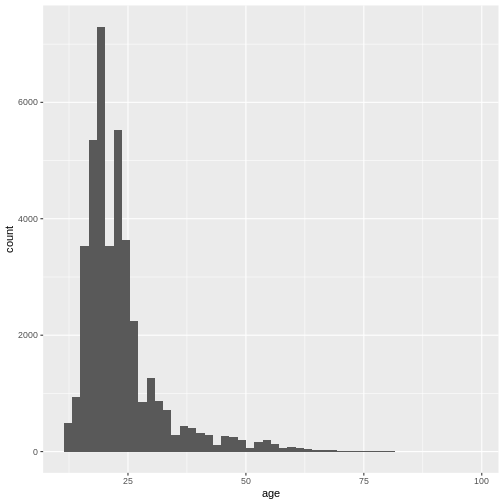
This makes it clear that valid_age_dass is the dataset
being used, and it improves readability.
Using the pipe for the second argument
By default, the pipe places the output of the previous code as the
first argument in the following function. In most cases, this is exactly
what we want to have. If you want to be more explicit about where the
pipe should place the output, you can do this by placing a
. as the placeholder for piped-data.
R
n_bins <- 30
n_bins %>%
hist(valid_age_dass$age, .)
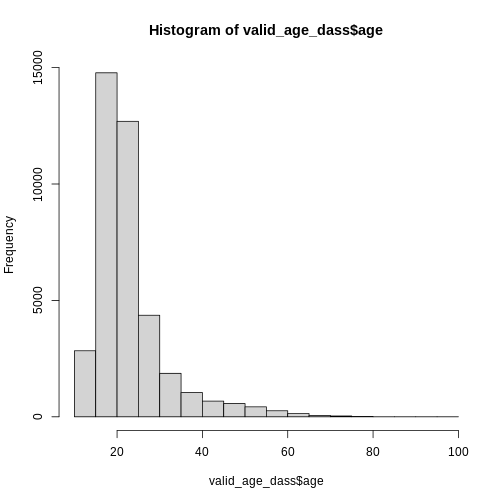
Numbered vs. Named Arguments
The pipe is made possible because most functions in the packages
included in tidyverse, like dplyr and
ggplot have the data as their first argument. This makes it
simple to pipe-through results from one function to the next, without
having to explicitly type function(data = something) every
time. In principle, almost all functions have a natural order in which
they expect arguments to occur. For example: ggplot()
expects data as the first argument and information about the mapping as
the second argument. This is why we always typed:
R
ggplot(
data = valid_age_dass,
mapping = aes(x = age)
)+
geom_density()
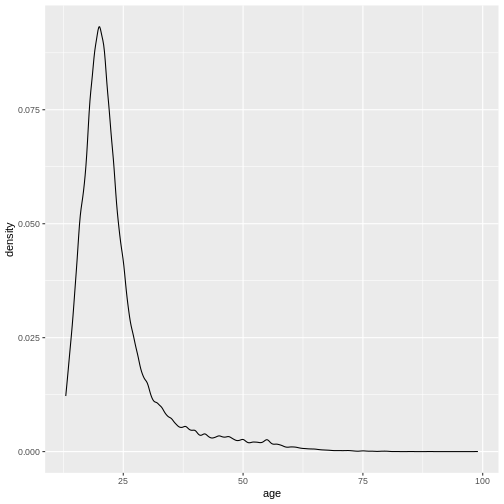
Notice that if we don’t specify the argument names, we still receive the same results. The function just uses the inputs you give it to match the arguments in the order in which they occur.
R
ggplot(
valid_age_dass, # Now it just automatically thinks this is data =
aes(x = age) # and this is mapping =
)+
geom_density()
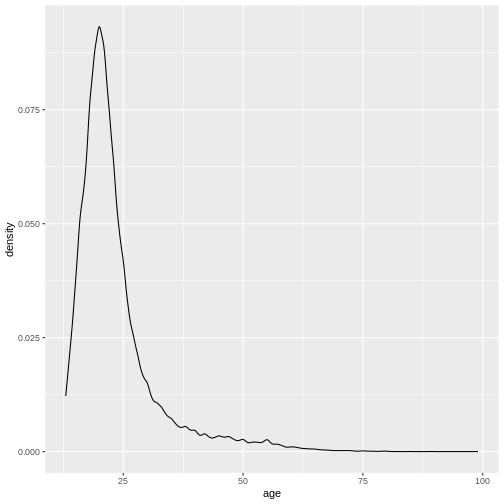
This is what makes this possible:
R
valid_age_dass %>%
ggplot(aes(x = age)) +
geom_density()
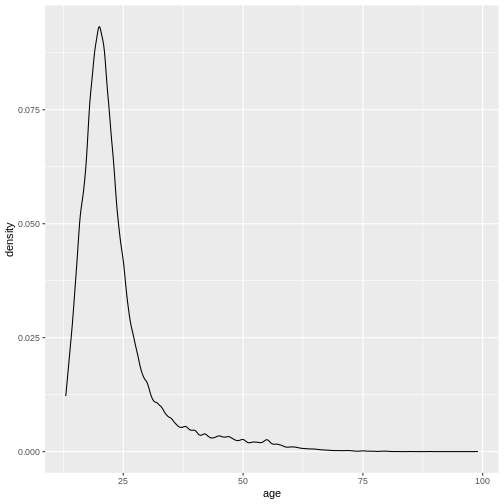
However, if we try to supply an argument in a place where it doesn’t belong, we get an error:
R
n_bins = 50
valid_age_dass %>%
ggplot(aes(x = age)) +
geom_histogram(n_bins) # This will cause an error!
ERROR
Error in `geom_histogram()`:
! `mapping` must be created by `aes()`.
✖ You've supplied a number.geom_histogram does not expect the number of bins to be
the first argument, but rather the mapping! Therefore, we need to
declare this properly:
R
valid_age_dass %>%
ggplot(aes(x = age)) +
geom_histogram(bins = n_bins)
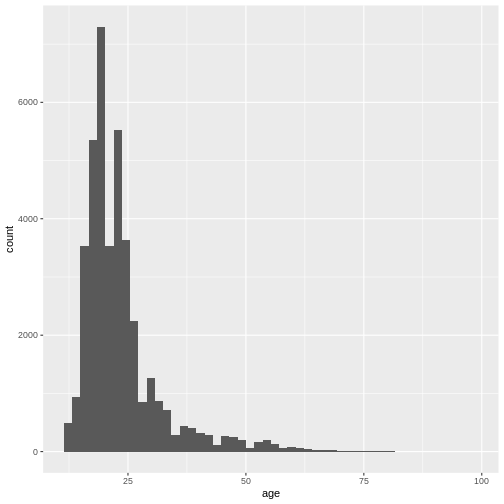
To learn about the order of arguments of a function, use
?function and inspect the help file.
Common Problems
Using one pipe too little
A common mistake is forgetting to use a pipe between arguments of a chain. For example:
R
valid_age_dass %>%
filter(age > 18)
select(age, country)
Here, select() is not part of the pipeline because the
pipe is missing after filter(age > 18). This can lead to
unexpected errors or incorrect results. The correct way to write this
is:
R
valid_age_dass %>%
filter(age > 18) %>%
select(age, country)
Using one pipe too much
Another common problem is using one pipe too many without having a function following it.
ERROR
Error in parse(text = input): <text>:4:0: unexpected end of input
2: filter(age > 18) %>%
3: select(age, country) %>%
^This can cause some really strange errors that never say anything about the pipe being superfluous. Make sure that all your pipe “lead somewhere”.
Using a pipe instead of + for
ggplot()
A source of confusion stems from the difference between the pipe
%>% and ggplot2’s +. They both
serve very similar purposes, namely adding together a chain of
functions. However, make sure you use %>% for everything
except ggplot-functions. Here, you will also receive an error
that is a little more informative:
R
valid_age_dass %>%
ggplot(aes(x = age)) %>%
geom_density()
ERROR
Error in `geom_density()` at magrittr/R/pipe.R:136:3:
! `mapping` must be created by `aes()`.
✖ You've supplied a <ggplot2::ggplot> object.
ℹ Did you use `%>%` or `|>` instead of `+`?Using = Instead of == in
filter()
Another common error is using = instead of
== when filtering data. For example:
R
valid_age_dass %>%
filter(country = "US") %>% # Incorrect!
head()
ERROR
Error in `filter()`:
! We detected a named input.
ℹ This usually means that you've used `=` instead of `==`.
ℹ Did you mean `country == "US"`?This will result in an error because = is used for
assigning values, while == is used for comparisons. The
correct syntax is:
R
valid_age_dass %>%
filter(country == "US") %>%
select(country, engnat, education) %>%
head()
OUTPUT
country engnat education
1 US 1 2
2 US 1 1
3 US 2 2
4 US 2 2
5 US 1 1
6 US 1 2Challenges
Challenge 1
Filter the complete dass_data. Create one dataset
containing only entries with people from the United States and one
dataset containing only entries from Great Britain. Compare the number
of entries in each dataset. What percentage of the total submissions
came from these two countries?
What is the average age of participants from the two countries?
Challenge 2
Create a filtered dataset based on the following conditions:
- The participant took less than 30 minutes to complete the DASS-part of the survey
- English is the native language of the participant
- There is an entry in the field
major
Challenge 3
What arguments does the cut() function take? Make a list
of the arguments and the order in which they have to be provided to the
function.
Challenge 4
In a single chain of functions, create a dataset is filtered with the same conditions as above and contains only the following variables:
- all demographic information, country information and the duration of the tests
- all the answers to the DASS (and only the answers, not the position or timing)
R
english_unversity_dass <- dass_data %>%
filter(testelapse < 1800) %>%
filter(engnat == 1) %>%
filter(major != "") %>%
select(starts_with("Q") & ends_with("A"), contains("elapse"), country, education:major)
write.csv(english_unversity_dass, "data/english_university_dass_data.csv")
Challenge 5
Save that dataset to a file
english_university_dass_data.csv in your
processed_data folder. You may need to use Google to find a
function that you can use to save data.
- Use the pipe operator
%>%to link multiple functions together - Use
filter()to filter rows based on certain conditions - Use
select()to keep only those rows that interest you
Content from Creating new columns
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What function is used to create new columns in a dataset?
- How can you generate a unique ID column for each row?
- How can you modify an existing column?
- How can you compute an average value over many columns in your data?
Objectives
- Understand how to use
mutate()to create new columns. - Learn how to add constant values, computed values, and conditionally assigned values to new columns.
- Use
ifelse()to create categorical columns based on conditions. - Learn efficient methods for computing row-wise sums and averages
using
rowSums()androwMeans(). - Understand the difference between
sum()androwSums()for column-wise and row-wise operations.
Making new columns
So far we have dealt with existing columns and worked on them. We
have computed descriptive statistics, plotted basic overviews of data
and cut the data just the way we like it using filter() and
select(). Now, we will unlock a new power… the power of
creation! Muhahahahaha.
Well, actually it’s just adding new columns to the dataset, but
that’s still cool! Let’s start by reading in our DASS data again and
loading the package dplyr.
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
To add a new column, we need the dplyr function
mutate(). It changes - or mutates - columns in the
dataset, like adding entirely new ones. The syntax is quite simple. The
first argument is the data, allowing easy use of pipes, and the second
argument is the code for the new column you want to have.
For example, if we want to create a new column that carries just a single value, we type the following:
R
dass_data <- dass_data %>%
mutate(single_value_column = 42)
To check if this worked:
R
unique(dass_data$single_value_column)
OUTPUT
[1] 42Saving progress
Notice how we had to assign the dataset to itself in order to save the progress we have made. If you do not do this, and just write down the pipeline, the end result will be outputted to the console, not stored in an object that we can use later.
R
dass_data %>%
head(6) %>%
mutate(some_column = "I have a value!") %>%
select(country, some_column)
OUTPUT
country some_column
1 IN I have a value!
2 US I have a value!
3 PL I have a value!
4 US I have a value!
5 MY I have a value!
6 US I have a value!You can easily assign the new data to something with the same name,
overriding the previous status. But be careful with this, I recommend
you use new names for the data after filtering, selecting, and mutating
existing columns. These operations change the original data (they remove
some information) and should thus be stored in a new object, like
filtered_dass_data.
We can do much more interesting things than just placing a constant
in the new column. For example, we can use row_number() to
create a running number of entries in our data - an id of sort.
R
dass_data <- dass_data %>%
mutate(entry_id = row_number())
dass_data %>%
select(country, testelapse, entry_id) %>%
head()
OUTPUT
country testelapse entry_id
1 IN 167 1
2 US 193 2
3 PL 271 3
4 US 261 4
5 MY 164 5
6 US 349 6Mutating existing columns
We can also base our mutation on existing columns.
testelapse is in seconds right now, let’s add a new column
that is the elapsed test time in minutes.
R
dass_data <- dass_data %>%
mutate(
testelapse_min = testelapse / 60
)
mean(dass_data$testelapse)
OUTPUT
[1] 2684.843R
mean(dass_data$testelapse_min)
OUTPUT
[1] 44.74738Another function I often use when creating new columns is
ifelse(). It works using three arguments.
R
a = 5
b = 3
ifelse(
a > b, # The condition
"a is greater", # IF yes
"a is smaller" # IF no
)
OUTPUT
[1] "a is greater"We can use this to make a new column that tells us whether an entry came from the United States.
R
dass_data <- dass_data %>%
mutate(is_from_us = ifelse(country == "US", "yes", "no"))
table(dass_data$is_from_us)
OUTPUT
no yes
31566 8207 Adding existing columns
One of the most important uses of mutate() is adding
together multiple columns. In our data, we have three columns indicating
the length of elapsed time during survey completion,
introelapse, testelapse and
surveyelapse.
We can compute the sum of these three columns to get the total amount of elapsed time.
R
dass_data <- dass_data %>%
mutate(
total_elapse_seconds = introelapse + testelapse + surveyelapse
)
Hint: Conduct Sanity Checks
When programming, you will make mistakes. There just is not a way around it. The only way to deal with it is trying to catch your mistakes as early as possible. Sometimes the computer will help you out by issuing an error or a warning. Sometimes it does not, because the code you wrote is technically valid but still does not exactly do what you want it to.
Therefore, it is important to conduct sanity checks! Try looking at the data and figure out if what you just did makes sense. In our case, let’s inspect the new column:
R
dass_data %>%
select(introelapse, testelapse, surveyelapse, total_elapse_seconds) %>%
head()
OUTPUT
introelapse testelapse surveyelapse total_elapse_seconds
1 19 167 166 352
2 1 193 186 380
3 5 271 122 398
4 3 261 336 600
5 1766 164 157 2087
6 4 349 213 566This looks good so far!
Now, this approach works well for a few columns, but gets tedious
quickly when attempting to compute the sum or average over multiple
columns. For this there exists a special fuction called
rowSums(). This function takes a dataset, and computes the
sums across all columns.
R
example_data <- data.frame(
x = 1:4,
y = 2:5,
z = 3:6
)
example_data$sum <- rowSums(example_data)
example_data
OUTPUT
x y z sum
1 1 2 3 6
2 2 3 4 9
3 3 4 5 12
4 4 5 6 15However, in our case we do not want a sum across all columns
of our data, but just some of the columns! To do this, we need to use
across() inside the rowSums function to select
specific columns.
R
dass_data %>%
mutate(elapse_sum = rowSums(across(c("introelapse", "testelapse", "surveyelapse")))) %>%
select(introelapse, testelapse, surveyelapse, elapse_sum) %>%
head()
OUTPUT
introelapse testelapse surveyelapse elapse_sum
1 19 167 166 352
2 1 193 186 380
3 5 271 122 398
4 3 261 336 600
5 1766 164 157 2087
6 4 349 213 566Because across() also accepts the same input as
select() we can use the special functions like
starts_with(), ends_with() or
contains().
R
dass_data %>%
mutate(elapse_sum = rowSums(across(ends_with("elapse")))) %>%
select(ends_with("elapse"), elapse_sum) %>%
head()
OUTPUT
introelapse testelapse surveyelapse elapse_sum
1 19 167 166 352
2 1 193 186 380
3 5 271 122 398
4 3 261 336 600
5 1766 164 157 2087
6 4 349 213 566Similarly, we can use rowMeans() to compute the average
test duration. The beautiful thing about mutate() is, that
we can create multiple columns in one function call!
R
dass_data %>%
mutate(
constant_value = "I am a value!",
id = row_number(),
elapse_sum = rowSums(across(ends_with("elapse"))),
elapse_mean = rowMeans(across(ends_with("elapse")))
) %>%
select(id, constant_value, ends_with("elapse"), elapse_sum, elapse_mean) %>%
head()
OUTPUT
id constant_value introelapse testelapse surveyelapse elapse_sum elapse_mean
1 1 I am a value! 19 167 166 352 117.3333
2 2 I am a value! 1 193 186 380 126.6667
3 3 I am a value! 5 271 122 398 132.6667
4 4 I am a value! 3 261 336 600 200.0000
5 5 I am a value! 1766 164 157 2087 695.6667
6 6 I am a value! 4 349 213 566 188.6667Pro Tip: mutate() + across()
Sometimes, you want to mutate multiple columns at once, applying the
same function to all of them. For example, when you want to recode some
values across all columns that share the incorrect format. Here, you can
use mutate() in combination with across() to
apply a function to all of them. In our case, lets compute the elapse
time in minutes for all elapse columns.
R
dass_data %>%
mutate(
across(
ends_with("elapse"), # Select the columns you want
~ .x / 60, # Write the function to apply to all of them
# Use this special function syntax ~ and a .x as the argument to be filled
.names = "{.col}_minutes" # Give out new names by using the old name
# which will take the place of {.col} and some new value.
)
) %>%
select(contains("elapse")) %>%
head()
OUTPUT
introelapse testelapse surveyelapse testelapse_min total_elapse_seconds
1 19 167 166 2.783333 352
2 1 193 186 3.216667 380
3 5 271 122 4.516667 398
4 3 261 336 4.350000 600
5 1766 164 157 2.733333 2087
6 4 349 213 5.816667 566
introelapse_minutes testelapse_minutes surveyelapse_minutes
1 0.31666667 2.783333 2.766667
2 0.01666667 3.216667 3.100000
3 0.08333333 4.516667 2.033333
4 0.05000000 4.350000 5.600000
5 29.43333333 2.733333 2.616667
6 0.06666667 5.816667 3.550000You don’t have to apply this straight away, and using this correctly takes quite a bit of practice. But I wanted you to know that this exists, so you might remember it when you need it.
Challenges
As you complete the challenges, make use of select() to
conduct sanity checks on the newly created columns!
Challenge 1
Create and save a new column called participant_id that includes a unique number per entry in the data.
Challenge 2
Create a new column that includes the total time the participant took to complete all surveys in minutes! Afterwards, filter this data to exclude all participants who took more than 1 hour. Work with this dataset for all following challenges.
Challenge 3
Create a column that indicates whether participants completed a university degree or not.
Challenge 4
Use a new function called case_when() to create a column
that has a translation from the numbers in the education
column to the verbal represenation. Use ?case_when() to
learn about this function. If that is not enough, try Google / ChatGPT.
Write a short description of what it does (1-2 sentences in comments of
script).
Challenge 5
The following code shows a common error when computing sums across columns.
R
# DO NOT USE THIS!
wrong_data <- dass_data %>%
mutate(wrong_elapse = sum(introelapse, testelapse, surveyelapse))
This code will execute without any errors and it looks correct. Can you figure out what went wrong here?
Challenge 6
Compute a column that contains the sum of all answers to the
DASS-questionnaire. Use rowSums() and
across().
R
dass_data <- dass_data %>%
mutate(
dass_score = rowSums(across(starts_with("Q") & ends_with("A")))
)
Challenge 7
Compute a column that shows the mean response to the DASS-questionnaire. Visualize the distribution across the dataset.
- Use
mutate()to create and modify columns in a dataset. - Assign constant values, compute values from other columns, or use conditions to define new columns.
- Use
ifelse()for conditional column creation. - Compute row-wise sums and means efficiently using
rowSums()androwMeans().
Content from Count and Summarize
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- How does
count()help in counting categorical data?
- How do you compute summary statistics?
- How can you use
group_by()to compute summary statistics for specific subgroups?
- Why is it useful to combine
count()withfilter()?
- How can you compute relative frequencies using
group_by()andmutate()?
Objectives
- Understand how to use
count()to summarize categorical variables.
- Learn to compute summary statistics using
summarize().
- Explore how
group_by()allows for subgroup analyses.
- Use
filter()to refine data before counting or summarizing.
- Compute and interpret relative frequencies using
mutate().
Computing Summaries
This lesson will teach you some of the last (and most important)
functions used in the process of data exploration and data analysis.
Today we will learn how to quickly count the number of appearances for a
given value using count() and how to compute custom summary
statistics using summarize().
Taken together, these functions will let you quickly compute frequencies and average values in your data, the last step in descriptive analysis and sometimes a necessary prerequisite for inference methods. The ANOVA, for example, most often requires one value per participant per condition, often requiring you to average over multiple trials.
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
Counting data
Let’s start with the more simple function of the two,
count(). This function works with the
tidyverse framework and can thus easily be used with pipes.
Simply pipe in your dataset and give the name of the column you want to
receive a count for:
R
dass_data %>%
count(education)
OUTPUT
education n
1 0 515
2 1 4066
3 2 15066
4 3 15120
5 4 5008The count will always be in a column called n, including
the number of occurrences for any given value (or combination of values)
to the left of it.
Notably, you can also include multiple columns to get a count for each combination of values.
R
dass_data %>%
count(hand, voted)
OUTPUT
hand voted n
1 0 0 4
2 0 1 51
3 0 2 118
4 1 0 284
5 1 1 9693
6 1 2 24765
7 2 0 36
8 2 1 1121
9 2 2 3014
10 3 0 3
11 3 1 183
12 3 2 503Tip: Using comparisons in
count()
You are not limited to comparing columns. You can also directly
include comparisons in the count() function. For example,
we can count the education status and number of people who concluded the
DASS-questionnaire in under 10 minutes.
R
dass_data %>%
count(testelapse < 600, education)
OUTPUT
testelapse < 600 education n
1 FALSE 0 33
2 FALSE 1 220
3 FALSE 2 1025
4 FALSE 3 922
5 FALSE 4 333
6 TRUE 0 482
7 TRUE 1 3846
8 TRUE 2 14041
9 TRUE 3 14198
10 TRUE 4 4675If you want to receive counts only for a specific part of the data,
like in our case people faster than 10 minutes, it may be more clear to
filter() first.
R
dass_data %>%
filter(testelapse < 600) %>%
count(education)
OUTPUT
education n
1 0 482
2 1 3846
3 2 14041
4 3 14198
5 4 4675Summarize
summarize() is one of the most powerful functions in
dplyr. Let’s use it to compute average scores for the
DASS-questionnaire.
First, compute columns with the sum DASS score and mean DASS score for each participant again. You should have code for this in the script from last episode. Go have a look and copy the correct code here. Copying from yourself is highly encouraged in coding! It’s very likely that you have spent some time solving a problem already, why not make use of that work.
Hint: Copy your own code
If you can’t find it or want to use the exact same specification as me, here is a solution.
R
dass_data <- dass_data %>%
mutate(
sum_dass_score = rowSums(across(starts_with("Q") & ends_with("A"))),
mean_dass_score = rowMeans(across(starts_with("Q") & ends_with("A")))
)
summarize() works similar to mutate() in
that you have to specify a new name and a formula to receive your
result. To get the average score of the DASS over all participants, we
use the mean() function.
R
dass_data %>%
summarize(
mean_sum_dass = mean(sum_dass_score)
)
OUTPUT
mean_sum_dass
1 100.2687We can also compute multiple means over both the column containing the sum of DASS scores and the mean DASS score.
R
dass_data %>%
summarize(
mean_sum_dass = mean(sum_dass_score),
mean_mean_dass = mean(mean_dass_score)
)
OUTPUT
mean_sum_dass mean_mean_dass
1 100.2687 2.387351In addition, we can compute other values using any functions we feel
are useful. For example, n() gives us the number of values,
which can be useful when comparing averages for multiple groups.
R
dass_data %>%
summarize(
mean_sum_dass = mean(sum_dass_score),
mean_mean_dass = mean(mean_dass_score),
n_subjecs = n(),
max_total_dass_score = max(sum_dass_score),
quantile_5_mean_dass = quantile(mean_dass_score, 0.05),
quantile_95_mean_dass = quantile(mean_dass_score, 0.95)
)
OUTPUT
mean_sum_dass mean_mean_dass n_subjecs max_total_dass_score
1 100.2687 2.387351 39775 168
quantile_5_mean_dass quantile_95_mean_dass
1 1.261905 3.595238Groups
So far, we could have done all of the above using basic R functions
like mean() or nrow(). One of the benefits of
summarize() is that it can be used in pipelines, so you
could have easily applied filters before summarizing. However, the most
important benefit is the ability to group the summary by other columns
in the data. In order to do this, you need to call
group_by().
This creates a special type of data format, called a grouped data
frame. R remembers assigns each row of the data a group based on
the values provided in the group_by() function. If you then
use summarize(), R will compute the summary functions for
each group separately.
R
dass_data %>%
group_by(education) %>%
summarize(
mean_mean_dass_score = mean(mean_dass_score),
n_subjects = n()
)
OUTPUT
# A tibble: 5 × 3
education mean_mean_dass_score n_subjects
<int> <dbl> <int>
1 0 2.38 515
2 1 2.64 4066
3 2 2.47 15066
4 3 2.31 15120
5 4 2.18 5008Now we are able to start learning interesting things about our data. For example, it seems like the average DASS score declines with higher levels of education (excluding the “missing” education = 0).
You can group by as many columns as you like. Summary statistics will then be computed for each combination of the columns:
R
dass_data %>%
group_by(education, urban) %>%
summarize(
mean_mean_dass_score = mean(mean_dass_score),
n_subjects = n()
)
OUTPUT
`summarise()` has grouped output by 'education'. You can override using the
`.groups` argument.OUTPUT
# A tibble: 20 × 4
# Groups: education [5]
education urban mean_mean_dass_score n_subjects
<int> <int> <dbl> <int>
1 0 0 2.31 19
2 0 1 2.48 126
3 0 2 2.35 141
4 0 3 2.35 229
5 1 0 2.66 49
6 1 1 2.69 626
7 1 2 2.59 1578
8 1 3 2.66 1813
9 2 0 2.62 156
10 2 1 2.46 3110
11 2 2 2.43 4720
12 2 3 2.50 7080
13 3 0 2.29 118
14 3 1 2.31 3351
15 3 2 2.28 5162
16 3 3 2.33 6489
17 4 0 2.18 40
18 4 1 2.19 1105
19 4 2 2.14 1631
20 4 3 2.20 2232Similar to count() you can also include comparisons in
group_by()
R
dass_data %>%
group_by(testelapse < 600) %>%
summarize(
mean_mean_dass_score = mean(mean_dass_score),
n_subjects = n()
)
OUTPUT
# A tibble: 2 × 3
`testelapse < 600` mean_mean_dass_score n_subjects
<lgl> <dbl> <int>
1 FALSE 2.33 2533
2 TRUE 2.39 37242Warning: Remember to
ungroup()
R remembers the group_by() function as long as you don’t
use ungroup() or summarize(). This may have
unwanted side-effects. Make sure that each group_by() is
followed by an ungroup().
R
dass_data %>%
group_by(education) %>%
mutate(
within_education_id = row_number()
) %>%
ungroup()
OUTPUT
# A tibble: 39,775 × 175
Q1A Q1I Q1E Q2A Q2I Q2E Q3A Q3I Q3E Q4A Q4I Q4E Q5A
<int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 4 28 3890 4 25 2122 2 16 1944 4 8 2044 4
2 4 2 8118 1 36 2890 2 35 4777 3 28 3090 4
3 3 7 5784 1 33 4373 4 41 3242 1 13 6470 4
4 2 23 5081 3 11 6837 2 37 5521 1 27 4556 3
5 2 36 3215 2 13 7731 3 5 4156 4 10 2802 4
6 1 18 6116 1 28 3193 2 2 12542 1 8 6150 3
7 1 20 4325 1 34 4009 2 38 3604 3 40 4826 4
8 1 34 4796 1 9 2618 1 39 5823 1 12 6596 3
9 4 4 3470 4 14 2139 3 1 11043 4 20 1829 3
10 3 38 5187 2 28 2600 4 9 2015 1 7 3111 4
# ℹ 39,765 more rows
# ℹ 162 more variables: Q5I <int>, Q5E <int>, Q6A <int>, Q6I <int>, Q6E <int>,
# Q7A <int>, Q7I <int>, Q7E <int>, Q8A <int>, Q8I <int>, Q8E <int>,
# Q9A <int>, Q9I <int>, Q9E <int>, Q10A <int>, Q10I <int>, Q10E <int>,
# Q11A <int>, Q11I <int>, Q11E <int>, Q12A <int>, Q12I <int>, Q12E <int>,
# Q13A <int>, Q13I <int>, Q13E <int>, Q14A <int>, Q14I <int>, Q14E <int>,
# Q15A <int>, Q15I <int>, Q15E <int>, Q16A <int>, Q16I <int>, Q16E <int>, …
count() + group_by()
You can also make use of group_by() to compute relative
frequencies in count-tables easily. For example, counting the education
status by handedness is not informative.
R
dass_data %>%
filter(hand %in% c(1, 2)) %>% # Focus on right vs. left
count(hand, education)
OUTPUT
hand education n
1 1 0 434
2 1 1 3481
3 1 2 13102
4 1 3 13326
5 1 4 4399
6 2 0 57
7 2 1 412
8 2 2 1623
9 2 3 1561
10 2 4 518This just tells us that there are more right-handed people. However, the percentage of education by handedness would be much more interesting. To that end, we can group by handedness and compute the percentage easily.
R
dass_data %>%
filter(hand %in% c(1, 2)) %>% # Focus on right vs. left
count(hand, education) %>%
group_by(hand) %>%
mutate(percentage = n / sum(n)) %>%
mutate(percentage = round(percentage*100, 2)) %>%
ungroup()
OUTPUT
# A tibble: 10 × 4
hand education n percentage
<int> <int> <int> <dbl>
1 1 0 434 1.25
2 1 1 3481 10.0
3 1 2 13102 37.7
4 1 3 13326 38.4
5 1 4 4399 12.7
6 2 0 57 1.37
7 2 1 412 9.88
8 2 2 1623 38.9
9 2 3 1561 37.4
10 2 4 518 12.4 As you can see, group_by() also works in combination
with mutate(). This means that calling sum(n)
computes the total of n for the given group. Which is why
n / sum(n) yields the relative frequency.
Challenges
Challenge 1
How many of the subjects voted in a national election last year? Use
count() to receive an overview.
Challenge 2
Get an overview of the number of people that voted based on whether their age is greater than 30. Make sure you only include subjects that are older than 18 in this count.
Challenge 3
Get a summary of the mean score on the DASS by whether somebody voted in a national election in past year. What do you notice? Why might this be the case? There are no right or wrong answers here, just think of some reasons.
Challenge 4
Get a summary of the mean DASS score based on age groups. Use
mutate() and case_when() to define a new
age_group column with the following cutoffs.
- < 18 = “underage”
- 18 - 25 = “youth”
- 26 - 35 = “adult”
- 36 - 50 = “middle-age”
- 50+ = “old”
R
dass_data %>%
mutate(
age_group = case_when(
age < 18 ~ "underage",
age >= 18 & age <= 25 ~ "youth",
age >= 26 & age <= 35 ~ "adult",
age >= 36 & age <= 50 ~ "middle-age",
age > 50 ~ "old"
)
) %>%
group_by(age_group) %>%
summarize(
mean_score = mean(mean_dass_score),
n_entries = n()
)
OUTPUT
# A tibble: 5 × 3
age_group mean_score n_entries
<chr> <dbl> <int>
1 adult 2.23 6229
2 middle-age 2.10 2288
3 old 1.97 950
4 underage 2.64 7269
5 youth 2.40 23039Challenge 5
Compute the relative frequency of whether somebody voted or not based on their age group (defined above). Make sure you compute the percentage of people that voted given their age, not the percentage of age given their voting status.
Challenge 6
Sort the above output by the relative frequency of people that voted.
Use the arrange() function after counting and computing the
percentage. Use the help ?arrange, Google or ChatGPT to
figure out how to use this function.
R
dass_data %>%
filter(voted > 0) %>% # Don't include missing values of voted = 0
mutate(
age_group = case_when(
age < 18 ~ "underage",
age >= 18 & age <= 25 ~ "youth",
age >= 26 & age <= 35 ~ "adult",
age >= 36 & age <= 50 ~ "middle-age",
age > 50 ~ "old"
)
) %>%
count(age_group, voted) %>%
group_by(age_group) %>%
mutate(
percentage = round(n / sum(n) * 100, 2)
) %>%
arrange(desc(percentage))
OUTPUT
# A tibble: 10 × 4
# Groups: age_group [5]
age_group voted n percentage
<chr> <int> <int> <dbl>
1 underage 2 7125 98.9
2 youth 2 17534 76.7
3 old 1 573 61.8
4 adult 1 3710 60.0
5 middle-age 1 1346 59.4
6 middle-age 2 919 40.6
7 adult 2 2468 40.0
8 old 2 354 38.2
9 youth 1 5337 23.3
10 underage 1 82 1.14Challenge 6 (continued)
What can you learn by the output?
- Use
count()to compute the number of occurrences for (combinations) of columns - Use
summarize()to compute any summary statistics for your data - Use
group_by()to group your data so you can receive summaries for each group separately - Combine functions like
filter(),group_by()andsummarize()using the pipe to receive specific results
Content from Midterms
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- No new questions today, only application
Objectives
- Apply learnings from previous episodes to new data
- Get to know data by visual inspection, data visualization and descriptive statistics
- Filter data to include only valid values
- Compute new columns
- Compute summary statistics
A new dataset approaches
Welcome to the “Midterms”! Today, we there will be no new lessons or functions that you have to understand. This episode is all about applying the things you already learnt to new data. We will use data collected by a previous Empra, who were investigating the impact of color on intelligence test performance.
Here, they tried to replicate an effect published by Elliot et al. (2007) concerning “the effect of red on performance attainment”. The original study showed that the presentation of the color red, for example by showing the participant a number in red, lowers the performance on a subsequent intelligence test. The tried to replicate this study online, by coloring the whole intelligence test red.
The performance of participants completing the test with red background could then be compared to participants with a green background and those with a grey/black background.
Additionally, a bachelor thesis integrated into the same project collected data on physical activity.
Your task
In this episode, there will only be challenges that contain the key functions introduced in earlier episodes. The challenges will guide you through the process of getting to know the data and applying your knowledge to new data. I will not provide solutions to this challenge in the current file, as you are encouraged to try solving this for yourself. As such, you may also use different methods than I, which lead to the same outcome or allow similar conclusions. If you get stuck, ask ChatGPT or a friend, or sleep on it! That often helped me.
Each challenge will include a short hint to the episode that contains information on how to solve it.
Try to write a “clean” script, using proper variable names and comments to describe what you are doing.
Challenges
Challenge 1 (Vectors and variable types)
Create a script called midterms_firstname_lastname.R in the
current project. Load the packages dplyr,
ggplot2 and psych. Install any packages that
you have not yet installed.
Challenge 2 (Projects)
Download the data here and store it in
your raw_data/ folder. Then load it into the current
session using an appropriate function.
Challenge 3 (Data Visualization (1))
Get to know the data. What are the column names? What type of information does each column store? Inspect the data visually and get the column names using an R function. Which columns are relevant for intelligence test performance? Which columns are related to physical activity?
Challenge 4 (Data Visualization (1) / Filtering data)
Get an overview of descriptive statistics of the whole dataset.
Inspect the mean, minimum and maximum values in the columns you think
are relevant. What do you notice? Use select() to create a
smaller dataset that only includes the columns you think are relevant to
make the overview simpler.
Challenge 5 (Filtering data)
The column item_kont includes information about the response to a control item. This item is so easy, that everyone should answer it correctly, given that they payed attention. If the value in item_kont is 1, the answer is correct. Otherwise it is incorrect.
Exclude all participants who did not answer correctly to the control item. Conduct sanity checks. How many participants were excluded? Did those participants have answers to the other questions?
Challenge 6 (Creating new columns)
Create new columns for each of the items that are named
itemNUMBER_accuracy. Each entry should be 1 if the
participant answered that item correctly and 0 if they did not.
If possible, use mutate() and across() for
this. Otherwise, simply use 12 lines of mutate(). Make sure
to conduct sanity checks whether this worked.
Challenge 7 (Creating new columns)
Create a column containing the total score of a participant. This should be the number of items they responded to correctly.
Challenge 8 (Data Visualization (2))
Compute key summary statistics of the total score of participants.
Inspect the distribution of values using hist(). What do
you notice? Are the minimum / maximum values plausible?
Challenge 9 (Filtering data)
Each item had 9 possible answers. Exclude participants who achieved a score that was below random chance.
Additionally, the columns beginning with dq_ include information about disqualification_criteria. Exclude participants based on the following criteria:
- dq_meaningless_responses = 3
- dq_distraction = 3
Challenge 10 (Count and summarize)
Count the number of occurrences of each of the three conditions in
fragebogen_der_im_interview_verwendet_wurde. Compute the
relative percentages.
Challenge 11 (Count and summarize)
Compute the average score by condition. Include the number of participants by condition and the standard deviation, too.
Challenge 12 - New!
Create a visualization showing the scores by condition. Here apply some of the knowledge from Data visualization (2) and add Google or ChatGPT to create a plot showing the condition differences (or lack thereof) in intelligence test scores.
- Apply what you have learned in new data!
Content from t-Test
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- When can I use a t-test?
- What type of t-test should I use?
- How can I run a t-test in R?
- How should I interpret the results of a t-test
Objectives
- Explain the assumptions behind a t-test.
- Demonstrate how to check the assumptions of a t-test.
- Show when to use a paired t-test, an independent samples t-test or a one-sample t-test.
- Learn practical applications of the t-test in R.
- Explain key issues in t-test interpretation.
Inferential statistics
So far, we have not “tested” anything. We have only described values and visualized distributions. We have stayed in the cozy realm of descriptive statistics. With descriptive statistics, we can describe data, we can look at a graph and say “this bar is higher than the other one”. Importantly, these conclusions are only truly valid for the present data! Meaning that we cannot generalize our results into other data, other samples, and more generally the population. Today, we will venture out into the different, scary world of inferential statistics that aims to address this problem.
Importantly, generalizing the results to a population is most often the goal of (psychological) research. Whatever a current sample does is irrelevant, we want to know whether the effect, the correlation, or the factor structure is present for the whole population from which we randomly selected some participants for our study.
This can be addressed by inferential (frequentist) statistics, asking the key question: “is this finding significant?”. Or, is my finding of an effect in a sample likely to be the result from a true effect in a population (significant) or just due to random sampling error.
Inferential statistics encompasses a large number of methods: t-tests, ANOVAs, linear regression only to name a few. For now, we will start out with the t-test which is useful when investigating mean values: Is the mean value of one group significantly higher than another group? Is my mean value significantly different from 0? This allows us to go beyond
Somehting about I will not go into details of statistical tests (recommendation).
Just broadly, we assume some distribution of the key statistic of interest (the test statistic) in the population given a null hypothesis (mean is 0, means are identical). This test statistic is… Then, we check our empirical test statistic. We can place this into the distribution to see how likely we are to observe this, given our assumptions in the test. In psychology we say that if it is less than 5% likely, we reject our assumptions of a null hypothesis.
This is true for all frequentists tests, they make some assumptions of a null hypothesis and the p-value indicates how likely we are to observe our current data given this null hypothesis. If we are less than 5% likely, we say that the null hypothesis is not true and reject it.
For example, something with mean value in dass data. Show some group differences based on education. Then illustrate the basic logic of the t-test.
show the basic t-test formula in r
then discuss assumptions of the t-test- - independence, random sampling, (approx) normal dist, variance - talk about robustness
show how to check the assumptions of the t-test. When is that an issue, when not?
There are different types of t-tests. Show what each of them are and show example uses in the dass data (maybe with half-splits in DASS?, check against two groups and criterium).
Now show how to interpret the t-test. Show and discuss the summary output. Explain how to report it in text and go over effect sizes!!
Show how to compute cohens d and why this is important.
Give personal recommendations. Focus on effect sizes, report p-values. Same when reading some paper. p-value is necessary, but it is mainly a function of sample size. So take with caution. Say something about no evidence for null. Something with BAYES
Now what if multiple groups? Outlook for next time: ANOVA
Challenge 1
Read in data and compute mean
Challenge 2
Check the assumptions of a t tesrt
Challenge 3
compute several t-tests, one-sample and independent samples
Challenge 4
compute t.test with different vectors and t-test using formula
Challenge 5
write a report of a t-test with effect size aswell.
- Something
Content from Factor Analysis - Introduction & EFA
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is a factor analysis?
- What is the difference between exploratory and confirmatory factor analysis?
- How can I run exploratory factor analysis in R?
- How can I interpret the results of an EFA in R?
Objectives
- Explain the idea behind factor analysis
- Understand the difference between exploratory and confirmatory factor analysis
- Learn how to conduct exploratory factor analysis in R
- Learn how to interpret the results of EFA in R
Factor analysis
Factor analysis is used to identify latent constructs—called factors—that explain the correlation patterns among observed variables. It helps reduce a large number of variables into fewer interpretable components.
It was first developed by Spearman in a field of study near to the heart of the author of this resource. Spearman noticed that students performing well in one subject in school tended to perform well in other subjects - there were high intercorrelations between academic performance. In order to investigate the structure behind these correlations, he used an early version of factor analysis to discover the now well known “g” factor of intelligence. One driving factor at the heart of intercorrelations of cognitive abilities.
This already illustrates several key principles of factor analysis. First, the key object of study are factors, mathematical constructs that can explain correlations between measured variables. In some fields such as structural equation modeling, these factors are sometimes referred to as latent variables. Latent, because they cannot be measured directly, contrary to manifest variables like academic performance. Importantly, these factors can show high validity in measuring a construct but are not the same! Construct validation requires a thorough research program before it can conclusively be stated that a certain factors is equal to a construct. For now, keep in mind that factors are mathematical in nature and can be a step towards measuring a construct, but do not necessarily do so.
Secondly, the key data in factor analysis are correlations! The underlying entry in any factor analysis is the covariance (or correlation) matrix. Modern statistical programs also allow us to provide the raw data and handle computing the correlation matrix, but these correlations are the core concept on which factor analysis is built.
Thirdly, there exists a difference between exploratory factor analysis (EFA) and confirmatory factor analysis (CFA).Exploratory Factor Analysis (EFA) is used when we do not have a predefined structure. It explores patterns and relationships. This is method is purely descriptive. In contrast Confirmatory Factor Analysis (CFA) is used when we do have a theory or structure to test. It evaluates how well the model fits the data. This is confirmatory and inferential, meaning we can test whether a given model fits our data, or doesn’t. Depending on whether you seek to explore the structure behind correlations of variables/items or test a hypothesized structure, choose the correct approach. Importantly, you cannot do both. You cannot use the same data to conduct an EFA, and then use this discovery in a subsequent CFA “testing” your factor structure. This needs to be done in an independent sample.
This is just a short introduction, details and further tutorial can be found here: https://lavaan.ugent.be/tutorial/cfa.html – CFA tutorial lavaan https://rpubs.com/pjmurphy/758265 – EFA tutorial Rpubs
Or in textbooks: - Stemmler et al (2016), Abschn. 2.1.4 - Tabachnik, B. G., & Fidell, L. S. (2001). Using multivariate statistics. 4th Ed. (chapter 13). Boston: Allyn and Bacon.
Examples using DASS data
Now, lets apply this theory to some actual data. Recall the DASS data from earlier lessons. Here, we have item-wise responses to a questionnaire claiming to measure three constructs, depression, anxiety, and stress
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
Let’s make our lives a little bit easier by only selecting the relevant columns for the subsequent factor analysis.
R
fa_data <- dass_data %>%
filter(testelapse < 600) %>%
select(starts_with("Q")& ends_with("A"))
psych::describe(fa_data)
OUTPUT
vars n mean sd median trimmed mad min max range skew kurtosis se
Q1A 1 37242 2.62 1.03 3 2.65 1.48 1 4 3 -0.03 -1.19 0.01
Q2A 2 37242 2.17 1.11 2 2.09 1.48 1 4 3 0.46 -1.16 0.01
Q3A 3 37242 2.23 1.04 2 2.16 1.48 1 4 3 0.40 -1.02 0.01
Q4A 4 37242 1.95 1.05 2 1.82 1.48 1 4 3 0.75 -0.71 0.01
Q5A 5 37242 2.53 1.07 2 2.54 1.48 1 4 3 0.04 -1.26 0.01
Q6A 6 37242 2.55 1.05 2 2.56 1.48 1 4 3 0.04 -1.21 0.01
Q7A 7 37242 1.93 1.04 2 1.79 1.48 1 4 3 0.79 -0.62 0.01
Q8A 8 37242 2.49 1.05 2 2.48 1.48 1 4 3 0.10 -1.20 0.01
Q9A 9 37242 2.67 1.07 3 2.71 1.48 1 4 3 -0.13 -1.26 0.01
Q10A 10 37242 2.46 1.14 2 2.45 1.48 1 4 3 0.10 -1.40 0.01
Q11A 11 37242 2.81 1.05 3 2.88 1.48 1 4 3 -0.28 -1.19 0.01
Q12A 12 37242 2.43 1.07 2 2.41 1.48 1 4 3 0.13 -1.22 0.01
Q13A 13 37242 2.79 1.07 3 2.86 1.48 1 4 3 -0.26 -1.26 0.01
Q14A 14 37242 2.58 1.08 2 2.61 1.48 1 4 3 -0.01 -1.29 0.01
Q15A 15 37242 1.83 0.99 2 1.67 1.48 1 4 3 0.94 -0.28 0.01
Q16A 16 37242 2.53 1.11 2 2.53 1.48 1 4 3 0.03 -1.35 0.01
Q17A 17 37242 2.66 1.16 3 2.71 1.48 1 4 3 -0.16 -1.44 0.01
Q18A 18 37242 2.47 1.07 2 2.46 1.48 1 4 3 0.10 -1.23 0.01
Q19A 19 37242 1.95 1.07 2 1.81 1.48 1 4 3 0.78 -0.74 0.01
Q20A 20 37242 2.33 1.12 2 2.29 1.48 1 4 3 0.25 -1.30 0.01
Q21A 21 37242 2.36 1.17 2 2.32 1.48 1 4 3 0.22 -1.43 0.01
Q22A 22 37242 2.35 1.03 2 2.31 1.48 1 4 3 0.25 -1.08 0.01
Q23A 23 37242 1.57 0.87 1 1.39 0.00 1 4 3 1.47 1.24 0.00
Q24A 24 37242 2.45 1.05 2 2.43 1.48 1 4 3 0.15 -1.18 0.01
Q25A 25 37242 2.19 1.08 2 2.11 1.48 1 4 3 0.42 -1.12 0.01
Q26A 26 37242 2.66 1.07 3 2.70 1.48 1 4 3 -0.12 -1.26 0.01
Q27A 27 37242 2.61 1.05 3 2.64 1.48 1 4 3 -0.04 -1.23 0.01
Q28A 28 37242 2.22 1.08 2 2.15 1.48 1 4 3 0.39 -1.13 0.01
Q29A 29 37242 2.65 1.06 3 2.69 1.48 1 4 3 -0.10 -1.24 0.01
Q30A 30 37242 2.40 1.08 2 2.37 1.48 1 4 3 0.17 -1.24 0.01
Q31A 31 37242 2.38 1.05 2 2.35 1.48 1 4 3 0.22 -1.13 0.01
Q32A 32 37242 2.45 1.02 2 2.44 1.48 1 4 3 0.15 -1.10 0.01
Q33A 33 37242 2.42 1.05 2 2.40 1.48 1 4 3 0.15 -1.18 0.01
Q34A 34 37242 2.64 1.15 3 2.67 1.48 1 4 3 -0.12 -1.44 0.01
Q35A 35 37242 2.31 1.00 2 2.26 1.48 1 4 3 0.31 -0.96 0.01
Q36A 36 37242 2.27 1.11 2 2.21 1.48 1 4 3 0.34 -1.24 0.01
Q37A 37 37242 2.38 1.14 2 2.35 1.48 1 4 3 0.21 -1.37 0.01
Q38A 38 37242 2.40 1.19 2 2.38 1.48 1 4 3 0.16 -1.49 0.01
Q39A 39 37242 2.45 1.02 2 2.44 1.48 1 4 3 0.15 -1.10 0.01
Q40A 40 37242 2.65 1.11 3 2.69 1.48 1 4 3 -0.13 -1.34 0.01
Q41A 41 37242 1.97 1.05 2 1.84 1.48 1 4 3 0.74 -0.72 0.01
Q42A 42 37242 2.69 1.03 3 2.74 1.48 1 4 3 -0.11 -1.20 0.01Beware of reversed items
This is especially relevant for EFA, but also for cfa as this might have some adverse impact
In theory, this questionnaire has a well-known proposed factor structure. It’s in the name - three factors: depression, anxiety, stress. Since it is also known which items are supposed to measure which factor, this would make the DASS data ideally suited for CFA, not EFA. However, for the sake of this exercise, let’s forget all of this for a second and pretend like we have no idea how the factor structure might look like.
As a first step, make sure the data is properly cleaned. After this is done, we can start the proper factor analysis process by investigating the correlation matrix of our data. Ideally, we will see high correlations between some items / variables. Should we observe no, or low correlations between all items, factor analysis will have a hard time identifying any underlying factors, as the items do not seem to share common influences.
Let’s start by computing the correlation matrix using
cor().
R
corr_mat <- cor(fa_data, use = "pairwise.complete.obs")
# head(corr_mat)
Now, we can visualize this using the package
ggcorrplot.
R
ggcorrplot::ggcorrplot(corr_mat)
WARNING
Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
ℹ Please use tidy evaluation idioms with `aes()`.
ℹ See also `vignette("ggplot2-in-packages")` for more information.
ℹ The deprecated feature was likely used in the ggcorrplot package.
Please report the issue at <https://github.com/kassambara/ggcorrplot/issues>.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.
In our case, we are greeted by a red square. A welcome sign for factor analysis as it indicates high correlations between items. This is even unusually uniform, other examples may show some variables correlating high with each other and low with other items. This is also perfectly normal. It’s only important that there are some correlations before proceeding with factor analysis.
Running the EFA
Now, we can already start running the EFA. In R, the easiest way to
do this is using fa.parallel() from the package
psych.
R
library(psych)
efa_result <- fa.parallel(fa_data, fa = "fa")
OUTPUT
Parallel analysis suggests that the number of factors = 9 and the number of components = NA Now, this function already gives you a lot of insight. First of all, you get two immediate results - a plot and a message informing you about the number of factors.
As a general rule, I would focus mainly on the plot and discard the message. This message is based on a resampling technique used to simulate new data without an underlying factor structure and then determines how many factors in the true data deviate significantly from this random simulation. In large datasets, this may overestimate the number of factors severly, as is the case in this example.
More important is the plot overall. It is called a Scree-plot and shows the eigenvalues of the factor solution. These eigenvalues indicate how much variance is explained by a factor. The larger the eigenvalue, the more variance explained by a factor.
These eigenvalues are already the basis of a selection technique on how many factors should be retained. Kaisers criterion states that all factors with eigenvalues > 1 should be extracted. In our case, these are the eigenvalues:
R
efa_result$fa.values
OUTPUT
[1] 18.588506243 2.318687720 1.042780083 0.543667379 0.425206073
[6] 0.257496536 0.145156657 0.094570513 0.041015178 0.028355531
[11] 0.001674441 -0.037595873 -0.041986500 -0.060701707 -0.065750863
[16] -0.082565635 -0.084817673 -0.095206394 -0.113248993 -0.115666333
[21] -0.129713392 -0.134490302 -0.144248475 -0.147258433 -0.150618100
[26] -0.153739659 -0.159396508 -0.166141135 -0.170279676 -0.172584141
[31] -0.175380247 -0.184334478 -0.188107697 -0.190790974 -0.194604989
[36] -0.195616247 -0.206315648 -0.243863018 -0.245374734 -0.248541704
[41] -0.280156397 -0.319514269In this case, 3 factors exceed eigenvalues of 1 and should thus be extracted (which aligns with theory nicely).
Another way of extracting the number of factors is by investigating the shape of the Scree plot directly. Here, we can see that the first factor has by far the largest value and thus already explains substantial variance. We can also see that all the other factors explain roughly the same amount of variance, with a possible exception of number 2 and 3. But certainly after the third factor, the amount of variance is low and decreases linearly.
In this method, we are looking to identify the “elbow” in the eigenvalue progression after which there is a sudden drop off in the variance explained. In this case, there is a strong argument that this occurs right after the first factor. The Scree plot would thus hint at only extracting one factor.
However, be cautious of relying on a single criterion to evaluate the number of factors. Kaisers criterion may be too liberal and the Scree plot may not be informative enough to warrant a certain guess. In the end, you should also select the number of factors that make the most sense psychologically and allow you to interpret the findings. You can not only rely on the eigenvalues to make this choice, but should also investigate the factor loadings of the items for a given number of factors.
Investigating Factor Loadings
Factor loadings can be determined for any given number of factors. They tell you how strongly the factor overall is related to a specific item. The higher, the better. The item with the highest loading on a factor is called the “marker item” and may inform you (in conjunction with the other items loading highly) as to what construct this factor actually reflects.
In principle, you are looking for simple structure. Items should load highly on one and only one factor while showing low loadings (< 0.3) on the other factors. If this is not the case, attributing an item to a single factor might not be warranted.
In order to obtain the factor loadings, we need to make two important choices. The first concerns the number of factors we want to investigate. For this example, let’s look at both one and three factors and investigate their structure. The second decision concerns the correlation between factors themselves. If you have strong reason to believe that the factors should be uncorrelated, or want to restrict the factors to be uncorrelated, i.e. independent of each other, you need to apply an orthoghonal rotation in the factor analyis. If you believe that the factors may themselves be related, you should apply an oblique rotation, which still allows for correlations between factors. In the case of only one extracted factor, this does not make a difference. In our specific example with three extracted factors, depression, anxiety, and stress may indeed be related. So we will proceed with the oblique rotation method called oblimin.
Let’s investigate the factor loadings for a three factor oblique solution.
R
fa_solution_3 <- psych::fa(fa_data, nfactors = 3, rotate = "oblimin")
OUTPUT
Loading required namespace: GPArotationR
print(fa_solution_3)
OUTPUT
Factor Analysis using method = minres
Call: psych::fa(r = fa_data, nfactors = 3, rotate = "oblimin")
Standardized loadings (pattern matrix) based upon correlation matrix
MR1 MR3 MR2 h2 u2 com
Q1A 0.08 0.73 -0.03 0.60 0.40 1.0
Q2A 0.05 0.07 0.42 0.26 0.74 1.1
Q3A 0.73 0.05 0.02 0.61 0.39 1.0
Q4A 0.03 -0.04 0.74 0.53 0.47 1.0
Q5A 0.62 0.13 0.04 0.56 0.44 1.1
Q6A -0.05 0.71 0.05 0.51 0.49 1.0
Q7A 0.01 -0.07 0.81 0.58 0.42 1.0
Q8A 0.16 0.37 0.27 0.50 0.50 2.3
Q9A -0.01 0.38 0.38 0.48 0.52 2.0
Q10A 0.89 -0.07 -0.02 0.69 0.31 1.0
Q11A 0.10 0.76 -0.07 0.62 0.38 1.1
Q12A -0.01 0.33 0.48 0.54 0.46 1.8
Q13A 0.65 0.19 0.02 0.66 0.34 1.2
Q14A -0.09 0.60 0.08 0.35 0.65 1.1
Q15A 0.14 -0.03 0.59 0.43 0.57 1.1
Q16A 0.77 0.03 -0.01 0.62 0.38 1.0
Q17A 0.74 0.07 0.01 0.64 0.36 1.0
Q18A 0.04 0.55 0.03 0.37 0.63 1.0
Q19A -0.01 0.03 0.58 0.35 0.65 1.0
Q20A 0.11 0.25 0.45 0.52 0.48 1.7
Q21A 0.88 -0.07 0.02 0.72 0.28 1.0
Q22A 0.17 0.41 0.20 0.49 0.51 1.8
Q23A 0.10 -0.05 0.58 0.37 0.63 1.1
Q24A 0.70 0.07 0.01 0.58 0.42 1.0
Q25A 0.00 0.02 0.66 0.46 0.54 1.0
Q26A 0.62 0.19 0.02 0.60 0.40 1.2
Q27A 0.11 0.70 -0.05 0.57 0.43 1.1
Q28A 0.00 0.24 0.58 0.58 0.42 1.3
Q29A 0.06 0.62 0.12 0.56 0.44 1.1
Q30A 0.13 0.34 0.27 0.43 0.57 2.2
Q31A 0.72 0.03 0.02 0.56 0.44 1.0
Q32A 0.04 0.60 0.06 0.45 0.55 1.0
Q33A 0.03 0.30 0.48 0.55 0.45 1.7
Q34A 0.76 0.07 0.00 0.66 0.34 1.0
Q35A 0.08 0.56 0.06 0.44 0.56 1.1
Q36A 0.18 0.24 0.40 0.53 0.47 2.1
Q37A 0.86 -0.08 0.00 0.64 0.36 1.0
Q38A 0.90 -0.09 0.01 0.70 0.30 1.0
Q39A 0.10 0.57 0.10 0.52 0.48 1.1
Q40A 0.02 0.37 0.36 0.47 0.53 2.0
Q41A 0.00 -0.07 0.77 0.53 0.47 1.0
Q42A 0.51 0.16 0.03 0.43 0.57 1.2
MR1 MR3 MR2
SS loadings 9.01 6.93 6.32
Proportion Var 0.21 0.17 0.15
Cumulative Var 0.21 0.38 0.53
Proportion Explained 0.40 0.31 0.28
Cumulative Proportion 0.40 0.72 1.00
With factor correlations of
MR1 MR3 MR2
MR1 1.00 0.70 0.59
MR3 0.70 1.00 0.69
MR2 0.59 0.69 1.00
Mean item complexity = 1.3
Test of the hypothesis that 3 factors are sufficient.
df null model = 861 with the objective function = 27.87 with Chi Square = 1037323
df of the model are 738 and the objective function was 2.08
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.03
The harmonic n.obs is 37242 with the empirical chi square 41215.05 with prob < 0
The total n.obs was 37242 with Likelihood Chi Square = 77517.62 with prob < 0
Tucker Lewis Index of factoring reliability = 0.914
RMSEA index = 0.053 and the 90 % confidence intervals are 0.053 0.053
BIC = 69750.03
Fit based upon off diagonal values = 1
Measures of factor score adequacy
MR1 MR3 MR2
Correlation of (regression) scores with factors 0.98 0.96 0.96
Multiple R square of scores with factors 0.96 0.93 0.92
Minimum correlation of possible factor scores 0.92 0.85 0.83Let’s take a closer look at the factor loadings:
R
print(fa_solution_3$loadings, cutoff = 0.3)
OUTPUT
Loadings:
MR1 MR3 MR2
Q1A 0.730
Q2A 0.423
Q3A 0.729
Q4A 0.741
Q5A 0.622
Q6A 0.714
Q7A 0.809
Q8A 0.366
Q9A 0.380 0.379
Q10A 0.886
Q11A 0.758
Q12A 0.325 0.477
Q13A 0.652
Q14A 0.598
Q15A 0.585
Q16A 0.771
Q17A 0.745
Q18A 0.553
Q19A 0.576
Q20A 0.446
Q21A 0.884
Q22A 0.412
Q23A 0.583
Q24A 0.705
Q25A 0.663
Q26A 0.616
Q27A 0.704
Q28A 0.580
Q29A 0.615
Q30A 0.344
Q31A 0.716
Q32A 0.604
Q33A 0.301 0.483
Q34A 0.760
Q35A 0.562
Q36A 0.405
Q37A 0.857
Q38A 0.895
Q39A 0.567
Q40A 0.368 0.364
Q41A 0.774
Q42A 0.509
MR1 MR3 MR2
SS loadings 8.037 5.409 5.109
Proportion Var 0.191 0.129 0.122
Cumulative Var 0.191 0.320 0.442Here, we can see that some items load highly one one of the three factors and do not load on any other factor (like 1-7). However, some items load on two factors (item 9, 12 etc). Here, we may want to investigate the items themselves to figure out which items load highly on which factor and why some of these items cannot be attributed to a single factor.
The marker items for the factors seem to be item 38 for factor 1, item 11 for factor 2, and item 7 for factor three. In the codebook, we can look up the item text itself.
- Item 38: “I felt that life was meaningless.”
- Item 11: “I found myself getting upset rather easily.”
- Item 7: “I had a feeling of shakiness (eg, legs going to give way).”
Now, one might see the factors Depression (meaningless life), Anxiety (getting upset) and Stress (shakiness) in these factors.
We can visualize the factor structure using
fa.diagram().
R
fa.diagram(fa_solution_3, simple = TRUE, cut = 0.3, digits = 2)
Lastly, we always have to investigate the correlations between the factors themselves. The plot already reveals substantial correlations of the three factors between 0.6 and 0.7.
R
fa_solution_3$score.cor
OUTPUT
[,1] [,2] [,3]
[1,] 1.0000000 0.7470426 0.6650608
[2,] 0.7470426 1.0000000 0.7935208
[3,] 0.6650608 0.7935208 1.0000000This indicates that there might be a higher order factor influencing all three factors. In the next episode, we can define a model testing whether this is the case or not.
Challenge 1
Read in data and answer the question: do we have a hypothesis as to how the structure should be?
Challenge 2
Run an EFA, how many factors would you extract using the Kaiser criterion, how many using the scree-plot
Challenge 3
Choose three factors and investigate the item loadings. Do we have simple structure?
Challenge 4
Choose one factor and investigate the item loadings
- Something
Content from Factor Analysis - CFA
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is a confirmatory factor analysis?
- How can I run CFA in R?
- How can I specify CFA models using lavaan?
- How can I interpret the results of a CFA and EFA in R?
Objectives
- Understand the difference between exploratory and confirmatory factor analysis
- Learn how to conduct confirmatory factor analysis in R
- Learn how to interpret the results of confirmatory factor analysis in R
Confirmatory Factor Analysis (CFA)
In the previous lesson, we approached the DASS as if we had no knowledge about the underlying factor structure. However, this is not really the case, since there is a specific theory proposing three interrelated factors. We can use confirmatory factor analysis to test whether this theory can adequately explain the item correlations found here.
Importantly, we not only need a theory of the number of factors, but also which items are supposed to load on which factor. This will be necessary when establishing our CFA model. Furthermore, we will test whether a one-factor model also is supported by the data and compare these two models, evaluating which one is better.
The main R package we will be using is lavaan. This
package is widely used for Structural Equation Modeling (SEM) in R. CFA
is a specific type of model in the general SEM framework. The lavaan
homepage provides a great tutorial on CFA, as well (https://lavaan.ugent.be/tutorial/cfa.html). For a more
detailed tutorial in SEM and mathematics behind CFA, please refer to the
lavaan homepage or this
introduction.
For now, let’s walk through the steps of a CFA in our specific case. First of all, in order to do confirmatory factor analysis, we need to have a hypothesis to confirm. How many factors, which variables are supposed to span which factor? Secondly, we need to investigate the correlation matrix for any issues (low correlations, correlations = 1). Then, we need to define our model properly using lavaan syntax. And finally, we can run our model and interpret our results.
Model Specification
Let’s start by defining the theory and the model syntax.
The DASS has 42 items and measures three scales: depression, anxiety, and stress. These scale are said to be intercorrelated, as they share common causes (genetic, environmental etc.). Furthermore, the DASS provides a detailed manual as to which items belong to which scales. The sequence of items and their respective scales is:
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
fa_data <- dass_data %>%
filter(testelapse < 600) %>%
select(starts_with("Q")& ends_with("A"))
item_scales_dass <- c(
"S", "A", "D", "A", "D", "S", "A",
"S", "A", "D", "S", "S", "D", "S",
"A", "D", "D", "S", "A", "A", "D",
"S", "A", "D", "A", "D", "S", "A",
"S", "A", "D", "S", "S", "D", "S",
"A", "D", "D", "S", "A", "A", "D"
)
item_scales_dass_data <- data.frame(
item_nr = 1:42,
scale = item_scales_dass
)
item_scales_dass_data
OUTPUT
item_nr scale
1 1 S
2 2 A
3 3 D
4 4 A
5 5 D
6 6 S
7 7 A
8 8 S
9 9 A
10 10 D
11 11 S
12 12 S
13 13 D
14 14 S
15 15 A
16 16 D
17 17 D
18 18 S
19 19 A
20 20 A
21 21 D
22 22 S
23 23 A
24 24 D
25 25 A
26 26 D
27 27 S
28 28 A
29 29 S
30 30 A
31 31 D
32 32 S
33 33 S
34 34 D
35 35 S
36 36 A
37 37 D
38 38 D
39 39 S
40 40 A
41 41 A
42 42 DScales vs. Factors
There is an important difference between scales and factors. Again, factors represent mathematical constructs discovered or constructed during factor analysis. Scales represent a collection of items bound together by the authors of the test and said to measure a certain construct. These two words cannot be used synonymously. Scales refer to item collections whereas factors refer to solutions of factor analysis. A scale can build a factor, in fact its items should span a common factor if the scale is constructed well. But a scale is not the same thing as a factor.
Now, in order to define our model we need to keep the distinction between manifest and latent variables in mind. Our manifest, measured variables are the items. The factors are the latent variable. We can also define which manifest variables contribute to which latent variables. In lavaan, this can be specified in the model code using a special syntax.
R
library(lavaan)
OUTPUT
This is lavaan 0.6-21
lavaan is FREE software! Please report any bugs.R
model_cfa_3facs <- c(
"
# Model Syntax
# =~ is like in linear regression
# left hand is latent factor, right hand manifest variables contributing
# Define which items load on which factor
depression =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A
anxiety =~ Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A
stress =~ Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Define correlations between factors using ~~
depression ~~ anxiety
depression ~~ stress
anxiety ~~ stress
"
)
Now, we can fit this model using cfa().
R
fit_cfa_3facs <- cfa(
model = model_cfa_3facs,
data = fa_data,
)
To inspect the model results, we use summary().
R
summary(fit_cfa_3facs, fit.measures = TRUE, standardize = TRUE)
OUTPUT
lavaan 0.6-21 ended normally after 46 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 87
Number of observations 37242
Model Test User Model:
Test statistic 107309.084
Degrees of freedom 816
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 1037764.006
Degrees of freedom 861
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.897
Tucker-Lewis Index (TLI) 0.892
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -1857160.022
Loglikelihood unrestricted model (H1) -1803505.480
Akaike (AIC) 3714494.044
Bayesian (BIC) 3715235.735
Sample-size adjusted Bayesian (SABIC) 3714959.249
Root Mean Square Error of Approximation:
RMSEA 0.059
90 Percent confidence interval - lower 0.059
90 Percent confidence interval - upper 0.059
P-value H_0: RMSEA <= 0.050 0.000
P-value H_0: RMSEA >= 0.080 0.000
Standardized Root Mean Square Residual:
SRMR 0.042
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
depression =~
Q3A 1.000 0.808 0.776
Q5A 0.983 0.006 155.298 0.000 0.794 0.742
Q10A 1.156 0.007 175.483 0.000 0.934 0.818
Q13A 1.077 0.006 173.188 0.000 0.870 0.810
Q16A 1.077 0.006 165.717 0.000 0.870 0.782
Q17A 1.158 0.007 172.607 0.000 0.936 0.807
Q21A 1.221 0.007 182.633 0.000 0.986 0.844
Q24A 0.987 0.006 159.408 0.000 0.797 0.758
Q26A 1.020 0.006 162.876 0.000 0.824 0.771
Q31A 0.966 0.006 156.243 0.000 0.780 0.746
Q34A 1.169 0.007 175.678 0.000 0.944 0.819
Q37A 1.118 0.007 168.220 0.000 0.903 0.791
Q38A 1.229 0.007 180.035 0.000 0.993 0.834
Q42A 0.826 0.006 131.716 0.000 0.668 0.646
anxiety =~
Q2A 1.000 0.563 0.506
Q4A 1.282 0.014 93.089 0.000 0.722 0.691
Q7A 1.303 0.014 94.229 0.000 0.734 0.707
Q9A 1.323 0.014 93.485 0.000 0.745 0.696
Q15A 1.116 0.013 89.050 0.000 0.629 0.636
Q19A 1.077 0.013 83.014 0.000 0.606 0.565
Q20A 1.473 0.015 96.526 0.000 0.830 0.742
Q23A 0.898 0.011 84.756 0.000 0.506 0.584
Q25A 1.244 0.014 89.922 0.000 0.701 0.647
Q28A 1.468 0.015 98.046 0.000 0.827 0.767
Q30A 1.260 0.014 90.684 0.000 0.710 0.657
Q36A 1.469 0.015 96.658 0.000 0.827 0.744
Q40A 1.374 0.015 93.607 0.000 0.774 0.698
Q41A 1.256 0.014 91.905 0.000 0.707 0.674
stress =~
Q1A 1.000 0.776 0.750
Q6A 0.943 0.007 137.700 0.000 0.732 0.695
Q8A 0.974 0.007 142.400 0.000 0.756 0.717
Q11A 1.030 0.007 152.388 0.000 0.799 0.761
Q12A 0.969 0.007 139.655 0.000 0.752 0.704
Q14A 0.797 0.007 111.328 0.000 0.619 0.572
Q18A 0.824 0.007 116.739 0.000 0.639 0.598
Q22A 0.947 0.007 141.490 0.000 0.735 0.713
Q27A 0.995 0.007 146.258 0.000 0.772 0.734
Q29A 1.020 0.007 149.022 0.000 0.791 0.746
Q32A 0.872 0.007 130.508 0.000 0.677 0.663
Q33A 0.970 0.007 142.051 0.000 0.753 0.715
Q35A 0.849 0.007 129.451 0.000 0.658 0.658
Q39A 0.956 0.007 144.729 0.000 0.742 0.727
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
depression ~~
anxiety 0.326 0.004 73.602 0.000 0.716 0.716
stress 0.491 0.005 93.990 0.000 0.784 0.784
anxiety ~~
stress 0.381 0.005 76.884 0.000 0.873 0.873
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.Q3A 0.431 0.003 128.109 0.000 0.431 0.398
.Q5A 0.515 0.004 129.707 0.000 0.515 0.450
.Q10A 0.431 0.003 125.297 0.000 0.431 0.331
.Q13A 0.398 0.003 125.958 0.000 0.398 0.345
.Q16A 0.481 0.004 127.782 0.000 0.481 0.389
.Q17A 0.467 0.004 126.117 0.000 0.467 0.348
.Q21A 0.394 0.003 122.826 0.000 0.394 0.288
.Q24A 0.471 0.004 129.017 0.000 0.471 0.426
.Q26A 0.463 0.004 128.367 0.000 0.463 0.405
.Q31A 0.486 0.004 129.556 0.000 0.486 0.444
.Q34A 0.439 0.004 125.238 0.000 0.439 0.330
.Q37A 0.487 0.004 127.220 0.000 0.487 0.374
.Q38A 0.430 0.003 123.805 0.000 0.430 0.304
.Q42A 0.622 0.005 132.512 0.000 0.622 0.583
.Q2A 0.922 0.007 133.250 0.000 0.922 0.744
.Q4A 0.572 0.004 127.940 0.000 0.572 0.523
.Q7A 0.539 0.004 127.113 0.000 0.539 0.500
.Q9A 0.590 0.005 127.667 0.000 0.590 0.515
.Q15A 0.581 0.004 130.109 0.000 0.581 0.595
.Q19A 0.785 0.006 132.085 0.000 0.785 0.681
.Q20A 0.561 0.004 124.984 0.000 0.561 0.449
.Q23A 0.493 0.004 131.617 0.000 0.493 0.658
.Q25A 0.681 0.005 129.719 0.000 0.681 0.581
.Q28A 0.478 0.004 123.083 0.000 0.478 0.412
.Q30A 0.662 0.005 129.348 0.000 0.662 0.568
.Q36A 0.551 0.004 124.837 0.000 0.551 0.446
.Q40A 0.631 0.005 127.580 0.000 0.631 0.513
.Q41A 0.601 0.005 128.683 0.000 0.601 0.546
.Q1A 0.467 0.004 126.294 0.000 0.467 0.437
.Q6A 0.571 0.004 129.082 0.000 0.571 0.516
.Q8A 0.541 0.004 128.138 0.000 0.541 0.486
.Q11A 0.464 0.004 125.600 0.000 0.464 0.421
.Q12A 0.574 0.004 128.705 0.000 0.574 0.504
.Q14A 0.785 0.006 132.627 0.000 0.785 0.672
.Q18A 0.733 0.006 132.077 0.000 0.733 0.642
.Q22A 0.523 0.004 128.331 0.000 0.523 0.492
.Q27A 0.510 0.004 127.255 0.000 0.510 0.461
.Q29A 0.498 0.004 126.551 0.000 0.498 0.443
.Q32A 0.585 0.004 130.300 0.000 0.585 0.561
.Q33A 0.541 0.004 128.213 0.000 0.541 0.489
.Q35A 0.568 0.004 130.460 0.000 0.568 0.567
.Q39A 0.491 0.004 127.618 0.000 0.491 0.471
depression 0.653 0.007 88.452 0.000 1.000 1.000
anxiety 0.317 0.006 50.766 0.000 1.000 1.000
stress 0.602 0.007 83.798 0.000 1.000 1.000Now, this output contains several key pieces of information:
- fit statistics for the model
- information on item loadings on the latent variables (factors)
- information on the correlations between the latent variables
- information on the variance of latent and manifest variables
Let’s walk through the output one-by-one.
Fit measures
The model fit is evaluated using three key criteria.
The first is the Chi^2 statistic. This evaluates whether the model adequately reproduces the covariance matrix in the real data or whether there is some deviation. Significant results indicate that the model does not reproduce the covariance matrix. However, this is largely dependent on sample size and not informative in practice. The CFA model is always a reduction of the underlying covariance matrix. That is the whole point of a model. Therefore, do not interpret the chi-square test and its associated p-value. Nonetheless, it is customary to report this information when presenting the model fit.
The following two fit statistics are independent of sample size and widely accepted in practice.
The CFI (Comparative Fit Index) is an indicator of fit. It ranges between 0 and 1 with 0 meaning horrible fit and 1 reflecting perfect fit. Values above 0.8 are acceptable and CFI > 0.9 is considered good. This index is independent of sample size and robust against violations of some assumptions CFA makes. It is however dependent on the number of free parameters as it does not include a penalty for additional parameters. Therefore, models with more parameter will generally have better CFI than parsimonious models with fewer parameters.
The RMSEA (Root Mean Squared Error Approximation) is an indicator of misfit that includes a penalty for the number of parameters. The higher the RMSEA, the worse. Generally, RMSEA < 0.05 is considered good and RMSEA < 0.08 acceptable. Models with RMSEA > 0.10 should not be interpreted. This fit statistic indicates whether the model fits the data well while controlling for the number of parameters specified.
It is customary to report all three of these measures when reporting the results of a CFA. Similarly, you should also interpret both the CFI and RMSEA, as bad fits in both statistics can indicate issues in the model specification. The above model would be reported as follows.
The model with three correlated factors showed acceptable fit to the data \(\chi^2(816) = 107309.08, p < 0.001, CFI = 0.90, RMSEA = 0.06\), 95% CI = \([0.06; 0.06]\).
In a future section, we will explore how you can compare models and select models based on these fit statistics. Here, it is important to focus on fit statistics that incorporate the number of free parameters like the RMSEA. Otherwise, more complex models will always outperform less complex models.
In addition to the statistics presented above, model comparison is often based on information criteria, like the AIC (Akaike Information Criterion) or the BIC (Bayesian Information Criterion). Here, lower values indicate better models. These information criteria also penalize complexity and are thus well suited for model comparison. More on this in the later section.
Loadings on latent variables
The next section in the model summary output display information on the latent variables. Specifically how highly the manifest variables “load” onto the latent variables they were assigned to. Here, loadings > 0.6 are considered high and at least one of the manifest variables should load highly on the latent variables. If all loadings are low, this might indicate that the factor you are trying to define does not really exist, as there is little relation between the manifest variables specified to load on this factor.
Importantly, the output shows two columns with loadings. The first column “Estimate” shows unstandardized loadings (like regression weights in a linear model). The last two columns “std.lv” and “std.all” show two different standardization techniques of the loadings. For now, focus on the “std.all” column. This can be interpreted in similar fashion as the factor loadings in the EFA.
In our example, these loadings are looking very good, especially for the depression scale. But for all scales, almost all items show standardized loadings of > 0.6, indicating that they reflect the underlying factor well.
Key Difference between EFA and CFA
These loadings highlight a key difference between CFA and a factor analysis with a fixed number of factors as conducted following an EFA.
In the EFA case, item cross-loadings are allowed. An item can load onto all factors. This is not the case in theory (unless explicitly stated). Here, the items only load onto the factor as specified in the model syntax. This is also why we obtain different loadings and correlations between factors in this CFA example compared to the three-factor FA from the previous episode.
Correlations between latent variables
The next section in the output informs us about the covariances between the latent variables. The “Estimate” column again shows the unstandardized solution while the “std.all” column reflects the standardized solution. The “std.all” column can be interpreted like a correlation (only on a latent level).
Here, we can see high correlations > 0.7 between our three factors. Anxiety and stress even correlate to 0.87! Maybe there is one underlying “g” factor of depression in our data after all? We will investigate this further in the section regarding model comparison.
Variances
The last section of the summary output displays the variances of the errors of manifest variables and the variance of the latent variables. Here, you should make sure that all values in “Estimate” are positive and that the variance of the latent variables differs significantly from 0, the results for this test are printed in “P(>|z|)”.
For CFA this section is not that relevant and plays a larger role in other SEM models.
Model Comparison
The last thing we will learn in this episode is how to leverage the fit statistics and other model information to compare different models. For example, remember that the EFA showed that the first factor had an extremely large eigenvalue, indicating that one factor already explained a substantial amount of variance.
A skeptic of the DASS might argue that it does not measure three distinct scales after all, but rather one unifying “I’m doing bad” construct. Therefore, this skeptic might generate a competing model of the DASS, with only one factor on which all items are loading.
Let’s first test the fit statistics of this model and then compare the two approaches.
R
model_cfa_1fac <- c(
"
# Model Syntax G Factor Model DASS
# Define only one general factor
g_dass =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A +Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A + Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
"
)
fit_cfa_1fac <- cfa(model_cfa_1fac, data = fa_data)
summary(fit_cfa_1fac, fit.measures = TRUE, standardize = TRUE)
OUTPUT
lavaan 0.6-21 ended normally after 25 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 84
Number of observations 37242
Model Test User Model:
Test statistic 235304.681
Degrees of freedom 819
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 1037764.006
Degrees of freedom 861
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.774
Tucker-Lewis Index (TLI) 0.762
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -1921157.820
Loglikelihood unrestricted model (H1) -1803505.480
Akaike (AIC) 3842483.641
Bayesian (BIC) 3843199.757
Sample-size adjusted Bayesian (SABIC) 3842932.805
Root Mean Square Error of Approximation:
RMSEA 0.088
90 Percent confidence interval - lower 0.087
90 Percent confidence interval - upper 0.088
P-value H_0: RMSEA <= 0.050 0.000
P-value H_0: RMSEA >= 0.080 1.000
Standardized Root Mean Square Residual:
SRMR 0.067
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
g_dass =~
Q3A 1.000 0.761 0.731
Q5A 1.016 0.007 142.019 0.000 0.773 0.722
Q10A 1.100 0.008 144.449 0.000 0.837 0.733
Q13A 1.113 0.007 156.040 0.000 0.846 0.788
Q16A 1.060 0.007 142.723 0.000 0.806 0.725
Q17A 1.147 0.008 148.618 0.000 0.872 0.753
Q21A 1.168 0.008 150.256 0.000 0.889 0.760
Q24A 0.991 0.007 140.991 0.000 0.754 0.717
Q26A 1.057 0.007 148.520 0.000 0.804 0.752
Q31A 0.962 0.007 137.412 0.000 0.732 0.700
Q34A 1.156 0.008 150.634 0.000 0.879 0.762
Q37A 1.063 0.008 139.320 0.000 0.809 0.709
Q38A 1.166 0.008 147.074 0.000 0.887 0.745
Q42A 0.865 0.007 124.363 0.000 0.658 0.637
Q2A 0.663 0.008 87.349 0.000 0.504 0.453
Q4A 0.806 0.007 114.105 0.000 0.613 0.587
Q7A 0.806 0.007 114.965 0.000 0.613 0.591
Q9A 0.901 0.007 125.202 0.000 0.686 0.641
Q15A 0.748 0.007 111.994 0.000 0.569 0.576
Q19A 0.675 0.007 92.306 0.000 0.513 0.478
Q20A 1.008 0.007 134.599 0.000 0.767 0.686
Q23A 0.585 0.006 99.515 0.000 0.445 0.514
Q25A 0.787 0.007 107.302 0.000 0.599 0.553
Q28A 0.968 0.007 134.071 0.000 0.737 0.684
Q30A 0.907 0.007 124.927 0.000 0.690 0.639
Q36A 1.034 0.007 139.134 0.000 0.787 0.708
Q40A 0.935 0.007 125.352 0.000 0.711 0.641
Q41A 0.773 0.007 108.798 0.000 0.588 0.560
Q1A 0.956 0.007 138.241 0.000 0.727 0.704
Q6A 0.869 0.007 122.694 0.000 0.661 0.629
Q8A 0.961 0.007 136.064 0.000 0.731 0.693
Q11A 0.987 0.007 140.627 0.000 0.751 0.715
Q12A 0.943 0.007 131.722 0.000 0.718 0.672
Q14A 0.724 0.007 98.622 0.000 0.551 0.510
Q18A 0.784 0.007 108.318 0.000 0.596 0.558
Q22A 0.929 0.007 134.440 0.000 0.707 0.685
Q27A 0.950 0.007 134.779 0.000 0.723 0.687
Q29A 0.972 0.007 137.073 0.000 0.740 0.698
Q32A 0.826 0.007 119.966 0.000 0.628 0.615
Q33A 0.954 0.007 135.255 0.000 0.725 0.689
Q35A 0.812 0.007 120.437 0.000 0.618 0.618
Q39A 0.916 0.007 133.922 0.000 0.697 0.683
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.Q3A 0.505 0.004 132.069 0.000 0.505 0.466
.Q5A 0.550 0.004 132.300 0.000 0.550 0.479
.Q10A 0.602 0.005 132.010 0.000 0.602 0.463
.Q13A 0.439 0.003 130.205 0.000 0.439 0.380
.Q16A 0.587 0.004 132.219 0.000 0.587 0.475
.Q17A 0.582 0.004 131.451 0.000 0.582 0.433
.Q21A 0.576 0.004 131.207 0.000 0.576 0.422
.Q24A 0.539 0.004 132.416 0.000 0.539 0.486
.Q26A 0.496 0.004 131.465 0.000 0.496 0.434
.Q31A 0.559 0.004 132.788 0.000 0.559 0.511
.Q34A 0.557 0.004 131.148 0.000 0.557 0.419
.Q37A 0.648 0.005 132.595 0.000 0.648 0.498
.Q38A 0.629 0.005 131.668 0.000 0.629 0.444
.Q42A 0.635 0.005 133.849 0.000 0.635 0.595
.Q2A 0.985 0.007 135.470 0.000 0.985 0.795
.Q4A 0.717 0.005 134.451 0.000 0.717 0.656
.Q7A 0.702 0.005 134.407 0.000 0.702 0.651
.Q9A 0.675 0.005 133.792 0.000 0.675 0.589
.Q15A 0.652 0.005 134.557 0.000 0.652 0.668
.Q19A 0.889 0.007 135.325 0.000 0.889 0.771
.Q20A 0.662 0.005 133.053 0.000 0.662 0.529
.Q23A 0.551 0.004 135.083 0.000 0.551 0.736
.Q25A 0.814 0.006 134.773 0.000 0.814 0.694
.Q28A 0.619 0.005 133.100 0.000 0.619 0.533
.Q30A 0.689 0.005 133.811 0.000 0.689 0.591
.Q36A 0.616 0.005 132.614 0.000 0.616 0.499
.Q40A 0.724 0.005 133.782 0.000 0.724 0.588
.Q41A 0.755 0.006 134.707 0.000 0.755 0.686
.Q1A 0.539 0.004 132.706 0.000 0.539 0.505
.Q6A 0.669 0.005 133.958 0.000 0.669 0.605
.Q8A 0.577 0.004 132.918 0.000 0.577 0.520
.Q11A 0.539 0.004 132.455 0.000 0.539 0.489
.Q12A 0.624 0.005 133.301 0.000 0.624 0.548
.Q14A 0.864 0.006 135.115 0.000 0.864 0.740
.Q18A 0.786 0.006 134.728 0.000 0.786 0.689
.Q22A 0.564 0.004 133.067 0.000 0.564 0.530
.Q27A 0.584 0.004 133.037 0.000 0.584 0.528
.Q29A 0.576 0.004 132.822 0.000 0.576 0.513
.Q32A 0.648 0.005 134.127 0.000 0.648 0.621
.Q33A 0.581 0.004 132.993 0.000 0.581 0.525
.Q35A 0.620 0.005 134.099 0.000 0.620 0.619
.Q39A 0.556 0.004 133.113 0.000 0.556 0.534
g_dass 0.579 0.007 81.612 0.000 1.000 1.000The one factor model showed a poor fit to the data \(\chi^2(819) = 235304.68, p < 0.001, CFI = 0.77, RMSEA = 0.09\), 95% CI = \([0.09; 0.09]\).
This already shows that the idea of only one unifying factor in the
DASS does not reflect the data well. We can formally test the difference
in the \(\chi^2\) statistic using
anova(), not to be confused with the ANOVA.
R
anova(fit_cfa_1fac, fit_cfa_3facs)
OUTPUT
Chi-Squared Difference Test
Df AIC BIC Chisq Chisq diff RMSEA Df diff Pr(>Chisq)
fit_cfa_3facs 816 3714494 3715236 107309
fit_cfa_1fac 819 3842484 3843200 235305 127996 1.0703 3 < 2.2e-16
fit_cfa_3facs
fit_cfa_1fac ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This output displays that the three factor model shows better information criteria (AIC and BIC) and that it also has lower (better) \(\chi^2\) statistics, significantly so. Coupled with an improved RMSEA for the three factor model, this evidence strongly suggests that the three factor model is better suited to explain the DASS data than the one factor model.
Challenge 1
Read in data and answer the question: do we have a hypothesis as to how the structure should be?
Challenge 2
Run a CFA with three factors
Challenge 3
Interpret some results
Challenge 4
Run a cfa with one factor, interpret some results
Challenge 5
Compare the models from challenge 3 and challenge 4, which one fits better?
- Something
Content from Factor Analysis - Advanced Measurement Models
Last updated on 2025-12-23 | Edit this page
Overview
Questions
- What is a bifactor model?
- What is a hierachical model?
- How can I specify different types of measurement models?
Objectives
- Understand the difference between hierarchical, non-hierarchical and bifactor models
- Learn how to specify a hierarchical model
- Learn how to specify a bifactor model
- Learn how to evaluate hierarchical and bifactor models
Measurement Models
So far, we have looked at only 1-factor and correlated multiple factor models in CFAs. This lesson, we will extend this approach to more general models. For example, CFA models can not only incorporate a number of factors correlating with each other, but also support a kind of “second CFA” on the levels of factors. If you have multiple factors that correlate highly with each other, they might themselves have a higher order factor that can account for their covariation.
This is called a “hierarchical” model. We can simply extend our three factor model as follows.
Hierarchical Models
R
library(lavaan)
OUTPUT
This is lavaan 0.6-21
lavaan is FREE software! Please report any bugs.R
model_cfa_hierarch <- c(
"
# Model Syntax
# Define which items load on which factor
depression =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A
anxiety =~ Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A
stress =~ Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Now, not correlation between factors, but hierarchy
high_level_fac =~ depression + anxiety + stress
"
)
Let’s load the data again and investigate this model:
R
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionR
dass_data <- read.csv("data/kaggle_dass/data.csv")
fa_data <- dass_data %>%
filter(testelapse < 600) %>%
select(starts_with("Q")& ends_with("A"))
fit_cfa_hierarch <- cfa(
model = model_cfa_hierarch,
data = fa_data,
)
summary(fit_cfa_hierarch, fit.measures = TRUE, standardized = TRUE)
OUTPUT
lavaan 0.6-21 ended normally after 38 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 87
Number of observations 37242
Model Test User Model:
Test statistic 107309.084
Degrees of freedom 816
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 1037764.006
Degrees of freedom 861
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.897
Tucker-Lewis Index (TLI) 0.892
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -1857160.022
Loglikelihood unrestricted model (H1) -1803505.480
Akaike (AIC) 3714494.044
Bayesian (BIC) 3715235.735
Sample-size adjusted Bayesian (SABIC) 3714959.249
Root Mean Square Error of Approximation:
RMSEA 0.059
90 Percent confidence interval - lower 0.059
90 Percent confidence interval - upper 0.059
P-value H_0: RMSEA <= 0.050 0.000
P-value H_0: RMSEA >= 0.080 0.000
Standardized Root Mean Square Residual:
SRMR 0.042
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
depression =~
Q3A 1.000 0.808 0.776
Q5A 0.983 0.006 155.298 0.000 0.794 0.742
Q10A 1.156 0.007 175.483 0.000 0.934 0.818
Q13A 1.077 0.006 173.188 0.000 0.870 0.810
Q16A 1.077 0.006 165.717 0.000 0.870 0.782
Q17A 1.158 0.007 172.607 0.000 0.936 0.807
Q21A 1.221 0.007 182.633 0.000 0.986 0.844
Q24A 0.987 0.006 159.408 0.000 0.797 0.758
Q26A 1.020 0.006 162.877 0.000 0.824 0.771
Q31A 0.966 0.006 156.243 0.000 0.780 0.746
Q34A 1.169 0.007 175.678 0.000 0.944 0.819
Q37A 1.118 0.007 168.220 0.000 0.903 0.791
Q38A 1.229 0.007 180.035 0.000 0.993 0.834
Q42A 0.826 0.006 131.716 0.000 0.668 0.646
anxiety =~
Q2A 1.000 0.563 0.506
Q4A 1.282 0.014 93.090 0.000 0.722 0.691
Q7A 1.303 0.014 94.229 0.000 0.734 0.707
Q9A 1.323 0.014 93.485 0.000 0.745 0.696
Q15A 1.116 0.013 89.051 0.000 0.629 0.636
Q19A 1.077 0.013 83.014 0.000 0.606 0.565
Q20A 1.473 0.015 96.526 0.000 0.830 0.742
Q23A 0.898 0.011 84.756 0.000 0.506 0.584
Q25A 1.244 0.014 89.922 0.000 0.701 0.647
Q28A 1.468 0.015 98.046 0.000 0.827 0.767
Q30A 1.260 0.014 90.684 0.000 0.710 0.657
Q36A 1.469 0.015 96.658 0.000 0.827 0.744
Q40A 1.374 0.015 93.607 0.000 0.774 0.698
Q41A 1.256 0.014 91.905 0.000 0.707 0.674
stress =~
Q1A 1.000 0.776 0.750
Q6A 0.943 0.007 137.700 0.000 0.732 0.695
Q8A 0.974 0.007 142.400 0.000 0.756 0.717
Q11A 1.030 0.007 152.388 0.000 0.799 0.761
Q12A 0.969 0.007 139.655 0.000 0.752 0.704
Q14A 0.797 0.007 111.328 0.000 0.619 0.572
Q18A 0.824 0.007 116.739 0.000 0.639 0.598
Q22A 0.947 0.007 141.491 0.000 0.735 0.713
Q27A 0.995 0.007 146.259 0.000 0.772 0.734
Q29A 1.020 0.007 149.023 0.000 0.791 0.746
Q32A 0.872 0.007 130.509 0.000 0.677 0.663
Q33A 0.970 0.007 142.052 0.000 0.753 0.715
Q35A 0.849 0.007 129.452 0.000 0.658 0.658
Q39A 0.956 0.007 144.730 0.000 0.742 0.727
high_level_fac =~
depression 1.000 0.802 0.802
anxiety 0.776 0.009 88.349 0.000 0.893 0.893
stress 1.171 0.009 124.409 0.000 0.978 0.978
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.Q3A 0.431 0.003 128.109 0.000 0.431 0.398
.Q5A 0.515 0.004 129.707 0.000 0.515 0.450
.Q10A 0.431 0.003 125.297 0.000 0.431 0.331
.Q13A 0.398 0.003 125.958 0.000 0.398 0.345
.Q16A 0.481 0.004 127.782 0.000 0.481 0.389
.Q17A 0.467 0.004 126.117 0.000 0.467 0.348
.Q21A 0.394 0.003 122.826 0.000 0.394 0.288
.Q24A 0.471 0.004 129.017 0.000 0.471 0.426
.Q26A 0.463 0.004 128.367 0.000 0.463 0.405
.Q31A 0.486 0.004 129.556 0.000 0.486 0.444
.Q34A 0.439 0.004 125.238 0.000 0.439 0.330
.Q37A 0.487 0.004 127.220 0.000 0.487 0.374
.Q38A 0.430 0.003 123.805 0.000 0.430 0.304
.Q42A 0.622 0.005 132.512 0.000 0.622 0.583
.Q2A 0.922 0.007 133.250 0.000 0.922 0.744
.Q4A 0.572 0.004 127.940 0.000 0.572 0.523
.Q7A 0.539 0.004 127.113 0.000 0.539 0.500
.Q9A 0.590 0.005 127.667 0.000 0.590 0.515
.Q15A 0.581 0.004 130.109 0.000 0.581 0.595
.Q19A 0.785 0.006 132.085 0.000 0.785 0.681
.Q20A 0.561 0.004 124.984 0.000 0.561 0.449
.Q23A 0.493 0.004 131.617 0.000 0.493 0.658
.Q25A 0.681 0.005 129.719 0.000 0.681 0.581
.Q28A 0.478 0.004 123.083 0.000 0.478 0.412
.Q30A 0.662 0.005 129.348 0.000 0.662 0.568
.Q36A 0.551 0.004 124.837 0.000 0.551 0.446
.Q40A 0.631 0.005 127.580 0.000 0.631 0.513
.Q41A 0.601 0.005 128.683 0.000 0.601 0.546
.Q1A 0.467 0.004 126.294 0.000 0.467 0.437
.Q6A 0.571 0.004 129.082 0.000 0.571 0.516
.Q8A 0.541 0.004 128.138 0.000 0.541 0.486
.Q11A 0.464 0.004 125.600 0.000 0.464 0.421
.Q12A 0.574 0.004 128.705 0.000 0.574 0.504
.Q14A 0.785 0.006 132.627 0.000 0.785 0.672
.Q18A 0.733 0.006 132.077 0.000 0.733 0.642
.Q22A 0.523 0.004 128.331 0.000 0.523 0.492
.Q27A 0.510 0.004 127.255 0.000 0.510 0.461
.Q29A 0.498 0.004 126.551 0.000 0.498 0.443
.Q32A 0.585 0.004 130.300 0.000 0.585 0.561
.Q33A 0.541 0.004 128.213 0.000 0.541 0.489
.Q35A 0.568 0.004 130.460 0.000 0.568 0.567
.Q39A 0.491 0.004 127.618 0.000 0.491 0.471
.depression 0.233 0.003 77.473 0.000 0.357 0.357
.anxiety 0.064 0.002 41.710 0.000 0.202 0.202
.stress 0.027 0.002 14.464 0.000 0.044 0.044
high_level_fac 0.420 0.006 70.881 0.000 1.000 1.000Again, let’s look at the fit indices first. The hierarchical model with three factors showed acceptable fit to the data \(\chi^2(816) = 107309.08, p < 0.001, CFI = 0.90, RMSEA = 0.06\), 95% CI = \([0.06; 0.06]\). Similarly, the loadings on the first-order factors (depression, anxiety, stress) also look good and we do not observe any issues in the variance.
If you have a good memory, you might notice that the fit statistics are identical to those in the correlated 3-factors model. This is because the higher order factor in this case only restructures the bivariate correlations into factor loadings. However, this is not true for all number of factors. In models with more than 3 first-order factors, establishing a second-order factor loading on all of them introduces more constraints than allowing the factors to correlate freely.
R
# Semplot here?
The hierarchical model allows us to investigate two very important things. Firstly, we can investigate the presence of a higher order factor. This is only the case if 1) the model fit is acceptable, 2) the factor loadings on this second-order factor are high enough and 3) the variance of the second-order factor is significantly different from 0. In this case, all three things are true, supporting the conclusion that a model with a higher order factor that contributes variance to all lower factors fits the data well.
Secondly, they are extremely useful in Structural Equation Models (SEM), as this higher-order factor can then itself be correlated to other outcomes, and allows researchers to investigate how much the general factor is associated with other items.
Bi-Factor Models
In this hierarchical case, we assume that the higher-order factor captures the shared variance of all first-order factors. So the higher-order factor only contributes variance to the observed variables indirectly through the first-order factors (depression etc.).
Bi-factor models change this behavior significantly. They do not impose a hierarchical structure but rather partition the variance in the observed variables into a general factor and specific factors. In the case of the DASS, a bifactor model would assert that each item is mostly determined by a general “I’m doing bad factor” and depending on the item context, depression, anxiety, or stress specifically contribute to the response on the specific item.
In contrast to a hierarchical model, this g-factor is directly influencing responses in the items and is not related to the specific factors. These models are often employed when researchers have good reason to believe that there are some specific factors unrelated to general response, as for example when using multiple tests measuring the same construct. One might introduce a general factor for construct and several specific factors capturing measurement specific variance (e.g. g factor for Openness, specific factors for self-rating vs. other-rating).
The model syntax displays this direct loading and the uncorrelated latent variables:
R
model_cfa_bifac <- c(
"
# Model Syntax
# Define which items load on which factor
depression =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A
anxiety =~ Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A
stress =~ Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Direct loading of the items on the general factor
g_dass =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A +Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A + Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Define correlations between factors using ~~
depression ~~ 0*anxiety
depression ~~ 0*stress
anxiety ~~ 0*stress
g_dass ~~ 0*depression
g_dass ~~ 0*stress
g_dass ~~ 0*anxiety
"
)
R
fit_cfa_bifac <- cfa(model_cfa_bifac, data = fa_data)
summary(fit_cfa_bifac, fit.measures = TRUE, standardized = TRUE)
OUTPUT
lavaan 0.6-21 ended normally after 90 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 126
Number of observations 37242
Model Test User Model:
Test statistic 72120.789
Degrees of freedom 777
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 1037764.006
Degrees of freedom 861
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.931
Tucker-Lewis Index (TLI) 0.924
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -1839565.874
Loglikelihood unrestricted model (H1) -1803505.480
Akaike (AIC) 3679383.748
Bayesian (BIC) 3680457.922
Sample-size adjusted Bayesian (SABIC) 3680057.494
Root Mean Square Error of Approximation:
RMSEA 0.050
90 Percent confidence interval - lower 0.049
90 Percent confidence interval - upper 0.050
P-value H_0: RMSEA <= 0.050 0.968
P-value H_0: RMSEA >= 0.080 0.000
Standardized Root Mean Square Residual:
SRMR 0.031
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
depression =~
Q3A 1.000 0.476 0.457
Q5A 0.838 0.011 77.509 0.000 0.399 0.373
Q10A 1.408 0.014 104.191 0.000 0.670 0.587
Q13A 0.930 0.010 88.624 0.000 0.443 0.412
Q16A 1.143 0.012 92.973 0.000 0.544 0.489
Q17A 1.242 0.013 97.712 0.000 0.591 0.510
Q21A 1.494 0.014 108.013 0.000 0.711 0.609
Q24A 0.960 0.011 85.644 0.000 0.457 0.434
Q26A 0.862 0.011 81.459 0.000 0.410 0.384
Q31A 0.967 0.011 85.275 0.000 0.461 0.440
Q34A 1.252 0.013 99.210 0.000 0.596 0.517
Q37A 1.380 0.014 101.533 0.000 0.657 0.576
Q38A 1.546 0.014 107.851 0.000 0.736 0.618
Q42A 0.655 0.011 61.266 0.000 0.312 0.302
anxiety =~
Q2A 1.000 0.288 0.258
Q4A 1.573 0.037 42.513 0.000 0.452 0.433
Q7A 1.940 0.044 44.187 0.000 0.558 0.538
Q9A 0.232 0.018 12.886 0.000 0.067 0.062
Q15A 1.207 0.030 39.724 0.000 0.347 0.351
Q19A 1.288 0.033 38.640 0.000 0.370 0.345
Q20A 0.442 0.020 22.556 0.000 0.127 0.114
Q23A 1.100 0.028 39.596 0.000 0.316 0.365
Q25A 1.342 0.034 40.033 0.000 0.386 0.357
Q28A 0.667 0.021 31.767 0.000 0.192 0.178
Q30A 0.170 0.019 9.086 0.000 0.049 0.045
Q36A 0.402 0.019 21.113 0.000 0.116 0.104
Q40A 0.292 0.019 15.254 0.000 0.084 0.076
Q41A 1.866 0.043 43.679 0.000 0.537 0.511
stress =~
Q1A 1.000 0.381 0.368
Q6A 0.790 0.016 49.154 0.000 0.301 0.286
Q8A -0.056 0.014 -4.124 0.000 -0.021 -0.020
Q11A 1.079 0.017 62.272 0.000 0.411 0.391
Q12A -0.463 0.015 -29.980 0.000 -0.176 -0.165
Q14A 0.800 0.018 44.430 0.000 0.304 0.282
Q18A 0.754 0.017 43.691 0.000 0.287 0.269
Q22A 0.187 0.013 13.917 0.000 0.071 0.069
Q27A 0.993 0.017 58.180 0.000 0.378 0.359
Q29A 0.605 0.014 42.099 0.000 0.230 0.217
Q32A 0.737 0.016 46.342 0.000 0.281 0.275
Q33A -0.525 0.016 -33.597 0.000 -0.200 -0.190
Q35A 0.680 0.015 44.095 0.000 0.259 0.259
Q39A 0.538 0.014 38.401 0.000 0.205 0.201
g_dass =~
Q3A 1.000 0.647 0.621
Q5A 1.058 0.008 127.074 0.000 0.685 0.639
Q10A 1.034 0.008 131.792 0.000 0.669 0.586
Q13A 1.158 0.008 140.588 0.000 0.749 0.697
Q16A 1.036 0.008 127.946 0.000 0.670 0.603
Q17A 1.128 0.008 135.443 0.000 0.730 0.630
Q21A 1.100 0.008 139.112 0.000 0.711 0.609
Q24A 0.996 0.008 126.030 0.000 0.644 0.612
Q26A 1.103 0.008 133.013 0.000 0.713 0.668
Q31A 0.959 0.008 122.467 0.000 0.620 0.592
Q34A 1.138 0.008 137.814 0.000 0.736 0.638
Q37A 0.996 0.008 125.672 0.000 0.644 0.565
Q38A 1.086 0.008 135.445 0.000 0.703 0.590
Q42A 0.914 0.008 110.771 0.000 0.592 0.572
Q2A 0.764 0.010 78.619 0.000 0.494 0.444
Q4A 0.956 0.010 100.570 0.000 0.618 0.592
Q7A 0.947 0.009 100.271 0.000 0.613 0.590
Q9A 1.152 0.010 114.816 0.000 0.745 0.696
Q15A 0.856 0.009 96.201 0.000 0.554 0.560
Q19A 0.802 0.009 84.718 0.000 0.519 0.483
Q20A 1.245 0.011 117.876 0.000 0.805 0.720
Q23A 0.667 0.008 86.982 0.000 0.431 0.498
Q25A 0.953 0.010 97.541 0.000 0.617 0.570
Q28A 1.238 0.010 120.648 0.000 0.800 0.743
Q30A 1.095 0.010 109.651 0.000 0.709 0.656
Q36A 1.247 0.011 118.553 0.000 0.807 0.726
Q40A 1.173 0.010 113.292 0.000 0.759 0.684
Q41A 0.910 0.009 96.144 0.000 0.589 0.561
Q1A 1.111 0.010 113.901 0.000 0.719 0.695
Q6A 1.058 0.010 108.405 0.000 0.684 0.651
Q8A 1.187 0.010 118.873 0.000 0.768 0.729
Q11A 1.139 0.010 114.602 0.000 0.737 0.702
Q12A 1.260 0.010 122.913 0.000 0.815 0.764
Q14A 0.871 0.010 90.195 0.000 0.563 0.521
Q18A 0.916 0.010 95.101 0.000 0.593 0.555
Q22A 1.118 0.010 115.483 0.000 0.723 0.702
Q27A 1.091 0.010 110.804 0.000 0.706 0.671
Q29A 1.171 0.010 116.850 0.000 0.757 0.715
Q32A 0.968 0.009 103.368 0.000 0.626 0.613
Q33A 1.271 0.010 124.894 0.000 0.822 0.781
Q35A 0.945 0.009 103.042 0.000 0.611 0.611
Q39A 1.091 0.010 113.992 0.000 0.706 0.692
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
depression ~~
anxiety 0.000 0.000 0.000
stress 0.000 0.000 0.000
anxiety ~~
stress 0.000 0.000 0.000
depression ~~
g_dass 0.000 0.000 0.000
stress ~~
g_dass 0.000 0.000 0.000
anxiety ~~
g_dass 0.000 0.000 0.000
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.Q3A 0.439 0.003 129.116 0.000 0.439 0.405
.Q5A 0.519 0.004 131.501 0.000 0.519 0.452
.Q10A 0.406 0.003 120.568 0.000 0.406 0.312
.Q13A 0.398 0.003 128.596 0.000 0.398 0.345
.Q16A 0.492 0.004 128.021 0.000 0.492 0.398
.Q17A 0.460 0.004 125.804 0.000 0.460 0.343
.Q21A 0.354 0.003 115.937 0.000 0.354 0.259
.Q24A 0.484 0.004 130.257 0.000 0.484 0.437
.Q26A 0.465 0.004 130.540 0.000 0.465 0.407
.Q31A 0.499 0.004 130.430 0.000 0.499 0.455
.Q34A 0.433 0.003 124.935 0.000 0.433 0.325
.Q37A 0.456 0.004 122.826 0.000 0.456 0.350
.Q38A 0.381 0.003 115.828 0.000 0.381 0.269
.Q42A 0.621 0.005 133.654 0.000 0.621 0.581
.Q2A 0.912 0.007 132.163 0.000 0.912 0.736
.Q4A 0.506 0.004 117.060 0.000 0.506 0.463
.Q7A 0.391 0.004 96.997 0.000 0.391 0.363
.Q9A 0.586 0.005 130.242 0.000 0.586 0.512
.Q15A 0.549 0.004 126.066 0.000 0.549 0.563
.Q19A 0.746 0.006 127.751 0.000 0.746 0.647
.Q20A 0.585 0.005 129.861 0.000 0.585 0.468
.Q23A 0.463 0.004 126.185 0.000 0.463 0.618
.Q25A 0.642 0.005 125.465 0.000 0.642 0.548
.Q28A 0.484 0.004 128.458 0.000 0.484 0.417
.Q30A 0.661 0.005 131.247 0.000 0.661 0.567
.Q36A 0.571 0.004 129.586 0.000 0.571 0.463
.Q40A 0.647 0.005 130.851 0.000 0.647 0.526
.Q41A 0.466 0.004 105.809 0.000 0.466 0.424
.Q1A 0.407 0.004 113.121 0.000 0.407 0.381
.Q6A 0.548 0.004 125.196 0.000 0.548 0.495
.Q8A 0.521 0.004 126.156 0.000 0.521 0.469
.Q11A 0.391 0.004 108.324 0.000 0.391 0.355
.Q12A 0.444 0.004 109.048 0.000 0.444 0.390
.Q14A 0.757 0.006 128.517 0.000 0.757 0.649
.Q18A 0.708 0.005 128.759 0.000 0.708 0.620
.Q22A 0.535 0.004 130.122 0.000 0.535 0.503
.Q27A 0.466 0.004 116.394 0.000 0.466 0.421
.Q29A 0.497 0.004 127.087 0.000 0.497 0.442
.Q32A 0.572 0.004 127.129 0.000 0.572 0.548
.Q33A 0.392 0.004 101.742 0.000 0.392 0.354
.Q35A 0.561 0.004 128.205 0.000 0.561 0.560
.Q39A 0.501 0.004 128.729 0.000 0.501 0.481
depression 0.227 0.004 56.333 0.000 1.000 1.000
anxiety 0.083 0.004 23.169 0.000 1.000 1.000
stress 0.145 0.004 38.869 0.000 1.000 1.000
g_dass 0.418 0.006 65.244 0.000 1.000 1.000This model shows good fit to the data \(\chi^2(777) = 72120.79, p < 0.001, CFI = 0.93, RMSEA = 0.05\), 95% CI = \([0.05; 0.05]\). We can additionally observe no issues in the loadings on the general factor. For the specific factors, there are items that do not seem to measure anything factor-specific in addition to the general factor, as they show loadings near zero on their specific factor.
From this model, we can learn two things. First, a general factor seems to be well supported by the data. Indicating that one could take all the items of the DASS scale and compute one general “distress” score that can be meaningfully interpreted. Second, there remains some specific variance in the items attributable to each scale. Together with the findings from the 3 factor model, this supports the interpretation that the DASS measures general distress and a small specific contribution of stress, anxiety and depression. However, some of the items load considerably more on general distress than on the specific factors.
Model Comparison - Nested models
Lets formally compare the four models we tested here to declare a “winner”, the model that fits our present data best. To do this, we will investigate the fit index RMSEA, the information criteria and conduct a chi-square differences test. The model with the best RMSEA and AIC/BIC and acceptable CFI will be crowned our winner.
Let’s recompute the one-factor and three-factor model.
R
model_cfa_1fac <- c(
"
# Model Syntax G Factor Model DASS
# Define only one general factor
g_dass =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A +Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A + Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
"
)
fit_cfa_1fac <- cfa(model_cfa_1fac, data = fa_data)
model_cfa_3facs <- c(
"
# Model Syntax
# Define which items load on which factor
depression =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A
anxiety =~ Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A
stress =~ Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Define correlations between factors using ~~
depression ~~ anxiety
depression ~~ stress
anxiety ~~ stress
"
)
fit_cfa_3facs <- cfa(
model = model_cfa_3facs,
data = fa_data,
)
Now, lets look at the fit indices.
R
fit_results <- data.frame(
c("1fac", "3fac", "hierarch", "bifac")
)
fit_results[, 2:6] <- 0
colnames(fit_results) <- c("model", "df", "chisq", "pvalue", "cfi", "rmsea")
fit_results[1, 2:6] <- fitmeasures(fit_cfa_1fac, c("df", "chisq", "pvalue", "cfi", "rmsea"))
fit_results[2, 2:6] <- fitmeasures(fit_cfa_3facs, c("df", "chisq", "pvalue", "cfi", "rmsea"))
fit_results[3, 2:6] <- fitmeasures(fit_cfa_hierarch, c("df", "chisq", "pvalue", "cfi", "rmsea"))
fit_results[4, 2:6] <- fitmeasures(fit_cfa_bifac, c("df", "chisq", "pvalue", "cfi", "rmsea"))
fit_results
OUTPUT
model df chisq pvalue cfi rmsea
1 1fac 819 235304.68 0 0.7738596 0.08767983
2 3fac 816 107309.08 0 0.8972970 0.05919692
3 hierarch 816 107309.08 0 0.8972970 0.05919692
4 bifac 777 72120.79 0 0.9311953 0.04965364In this comparison, the bifactor model clearly winds out. It shows
the lowest RMSEA despite having more free parameters and shows good CFI.
Using anova(), we can formally test this.
R
anova(fit_cfa_1fac, fit_cfa_3facs, fit_cfa_hierarch, fit_cfa_bifac)
WARNING
Warning: lavaan->lavTestLRT():
some models have the same degrees of freedomOUTPUT
Chi-Squared Difference Test
Df AIC BIC Chisq Chisq diff RMSEA Df diff
fit_cfa_bifac 777 3679384 3680458 72121
fit_cfa_3facs 816 3714494 3715236 107309 35188 0.15556 39
fit_cfa_hierarch 816 3714494 3715236 107309 0 0.00000 0
fit_cfa_1fac 819 3842484 3843200 235305 127996 1.07032 3
Pr(>Chisq)
fit_cfa_bifac
fit_cfa_3facs < 2.2e-16 ***
fit_cfa_hierarch
fit_cfa_1fac < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Important - CFA vs. EFA
Important note, do not confuse confirmatory with exploratory research! You cannot use the same data and first run an EFA, then define some models and run a CFA to confirm your “best” model. It should be clearly stated then that this is exploratory research only. True model difference tests to confirm hypothesis can only be conducted if these hypothesis allow specification of the model before the data is seen!
A look to SEM
What we have investigated so far is also called a measurement model, because it allows the specification of latent factors that are supposed to measure certain constructs. In the broader context of Structural Equation Modelling (SEM), we can extend this CFA measurement model by combining multiple measurement models and investigating the relationship between latent factors of different measurement models in a regression model.
For example, we can use the bifactor model as a measurement model for general distress and then relate this factor to the measures of the big 5.
R
big5_distress_model <- c(
"
# Model Syntax
# Define which items load on which factor
depression =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A
anxiety =~ Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A
stress =~ Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Direct loading of the items on the general factor
g_dass =~ Q3A + Q5A + Q10A + Q13A + Q16A + Q17A + Q21A + Q24A + Q26A + Q31A + Q34A + Q37A + Q38A + Q42A +Q2A + Q4A + Q7A + Q9A + Q15A + Q19A + Q20A + Q23A + Q25A + Q28A + Q30A + Q36A + Q40A + Q41A + Q1A + Q6A + Q8A + Q11A + Q12A + Q14A + Q18A + Q22A + Q27A + Q29A + Q32A + Q33A + Q35A + Q39A
# Define correlations between factors using ~~
depression ~~ 0*anxiety
depression ~~ 0*stress
anxiety ~~ 0*stress
g_dass ~~ 0*depression
g_dass ~~ 0*stress
g_dass ~~ 0*anxiety
"
)
- Something
